Phylogenetic characterization and in situ detection of a Cytophaga-Flexibacter-Bacteroides phylogroup bacterium in Tuber borchii vittad. Ectomycorrhizal mycelium
- PMID: 11055961
- PMCID: PMC92417
- DOI: 10.1128/AEM.66.11.5035-5042.2000
Phylogenetic characterization and in situ detection of a Cytophaga-Flexibacter-Bacteroides phylogroup bacterium in Tuber borchii vittad. Ectomycorrhizal mycelium
Abstract
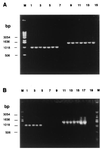
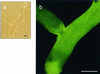
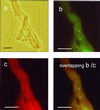
Mycorrhizal ascomycetous fungi are obligate ectosymbionts that colonize the roots of gymnosperms and angiosperms. In this paper we describe a straightforward approach in which a combination of morphological and molecular methods was used to survey the presence of potentially endo- and epiphytic bacteria associated with the ascomycetous ectomycorrhizal fungus Tuber borchii Vittad. Universal eubacterial primers specific for the 5' and 3' ends of the 16S rRNA gene (16S rDNA) were used for PCR amplification, direct sequencing, and phylogenetic analyses. The 16S rDNA was amplified directly from four pure cultures of T. borchii Vittad. mycelium. A nearly full-length sequence of the gene coding for the prokaryotic small-subunit rRNA was obtained from each T. borchii mycelium studied. The 16S rDNA sequences were almost identical (98 to 99% similarity), and phylogenetic analysis placed them in a single unique rRNA branch belonging to the Cytophaga-Flexibacter-Bacteroides (CFB) phylogroup which had not been described previously. In situ detection of the CFB bacterium in the hyphal tissue of the fungus T. borchii was carried out by using 16S rRNA-targeted oligonucleotide probes for the eubacterial domain and the Cytophaga-Flexibacter phylum, as well as a probe specifically designed for the detection of this mycelium-associated bacterium. Fluorescent in situ hybridization showed that all three of the probes used bound to the mycelium tissue. This study provides the first direct visual evidence of a not-yet-cultured CFB bacterium associated with a mycorrhizal fungus of the genus Tuber.
Figures

Similar articles
-
Competitive PCR for quantitation of a Cytophaga-Flexibacter-Bacteroides phylum bacterium associated with the Tuber borchii Vittad. mycelium.Appl Environ Microbiol. 2002 Dec;68(12):6421-4. doi: 10.1128/AEM.68.12.6421-6424.2002. Appl Environ Microbiol. 2002. PMID: 12450871 Free PMC article.
-
New degenerate Cytophaga-Flexibacter-Bacteroides-specific 16S ribosomal DNA-targeted oligonucleotide probes reveal high bacterial diversity in River Taff epilithon.Appl Environ Microbiol. 2002 Jan;68(1):201-10. doi: 10.1128/AEM.68.1.201-210.2002. Appl Environ Microbiol. 2002. PMID: 11772628 Free PMC article.
-
16S rRNA-targeted oligonucleotide probes for the in situ detection of members of the phylum Cytophaga-Flavobacterium-Bacteroides.Syst Appl Microbiol. 2000 Apr;23(1):107-14. doi: 10.1016/S0723-2020(00)80051-X. Syst Appl Microbiol. 2000. PMID: 10879984
-
Phylogenetic position of Chitinophaga pinensis in the Flexibacter-Bacteroides-Cytophaga phylum.Int J Syst Bacteriol. 1999 Apr;49 Pt 2:479-81. doi: 10.1099/00207713-49-2-479. Int J Syst Bacteriol. 1999. PMID: 10319467
-
Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment.Microbiology (Reading). 1996 May;142 ( Pt 5):1097-1106. doi: 10.1099/13500872-142-5-1097. Microbiology (Reading). 1996. PMID: 8704951
Cited by
-
Competitive PCR for quantitation of a Cytophaga-Flexibacter-Bacteroides phylum bacterium associated with the Tuber borchii Vittad. mycelium.Appl Environ Microbiol. 2002 Dec;68(12):6421-4. doi: 10.1128/AEM.68.12.6421-6424.2002. Appl Environ Microbiol. 2002. PMID: 12450871 Free PMC article.
-
Endohyphal bacterium enhances production of indole-3-acetic acid by a foliar fungal endophyte.PLoS One. 2013 Sep 24;8(9):e73132. doi: 10.1371/journal.pone.0073132. eCollection 2013. PLoS One. 2013. PMID: 24086270 Free PMC article.
-
The Role of the Microbiome of Truffles in Aroma Formation: a Meta-Analysis Approach.Appl Environ Microbiol. 2015 Oct;81(20):6946-52. doi: 10.1128/AEM.01098-15. Epub 2015 Jul 17. Appl Environ Microbiol. 2015. PMID: 26187969 Free PMC article. Review.
-
A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps.Proc Natl Acad Sci U S A. 2001 Oct 23;98(22):12555-60. doi: 10.1073/pnas.221467498. Epub 2001 Oct 9. Proc Natl Acad Sci U S A. 2001. PMID: 11592990 Free PMC article.
-
Diversity, Bacterial Symbionts, and Antimicrobial Potential of Termite-Associated Fungi.Front Microbiol. 2020 Mar 13;11:300. doi: 10.3389/fmicb.2020.00300. eCollection 2020. Front Microbiol. 2020. PMID: 32231643 Free PMC article.
References
-
- Bedini S, Bagnoli G, Sbrana C, Leporini C, Tola E, Dunne C, Filippi C, D'Andrea F, O'Gara F, Nuti M P. Pseudomonads isolated from within fruit bodies of T. borchii are capable of producing biological control or phytostimulatory compounds in pure culture. Symbiosis. 1999;26:223–236.
-
- Bernadet J F, Segers P, Vancanneyt M, Berthe F, Kesters K, Vandamme P. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga aquatilis Strohl and Tait 1978) Int J Syst Bacteriol. 1996;46:128–148.
-
- Bertini L, Agostini D, Potenza L, Rossi I, Zeppa S, Zambonelli A, Stocchi V. Molecular markers for the identification of the ectomycorrhizal fungus Tuber borchii. New Phytol. 1998;139:565–570.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

