Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo
- PMID: 11058103
- PMCID: PMC113125
- DOI: 10.1093/nar/28.21.4083
Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo
Abstract
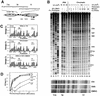
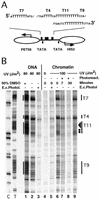
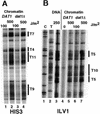
Poly(dA.dT) sequences (T-tracts) are abundant genomic DNA elements with unusual properties in vitro and an established role in transcriptional regulation of yeast genes. In vitro T-tracts are rigid, contribute to DNA bending, affect assembly in nucleosomes and generate a characteristic pattern of CPDs (cyclobutane pyrimidine dimers) upon irradiation with UV light (UV photofootprint). In eukaryotic cells, where DNA is packaged in chromatin, the DNA structure of T-tracts is unknown. Here we have used in vivo UV photofootprinting and DNA repair by photolyase to investigate the structure and accessibility of T-tracts in yeast promoters (HIS3, URA3 and ILV1). The same characteristic photofootprints were obtained in yeast and in naked DNA, demonstrating that the unusual T-tract structure exists in living cells. Rapid repair of CPDs in the T-tracts demonstrates that these T-tracts were not folded in nucleosomes. Moreover, neither datin, a T-tract binding protein, nor Gcn5p, a histone acetyltransferase involved in nucleosome remodelling, showed an influence on the structure and accessibility of T-tracts. The data support a contribution of this unusual DNA structure to transcriptional regulation.
Figures

References
-
- Nelson H.C., Finch,J.T., Luisi,B.F. and Klug,A. (1987) Nature, 330, 221–226. - PubMed
-
- Leroy J.L., Charretier,E., Kochoyan,M. and Gueron,M. (1988) Biochemistry, 27, 8894–8898. - PubMed
-
- Alexeev D.G., Lipanov,A.A. and Skuratovskii,I. (1987) Nature, 325, 821–823. - PubMed
-
- Peck L.J. and Wang,J.C. (1981) Nature, 292, 375–378. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

