A family of developmentally excised DNA elements in Tetrahymena is under selective pressure to maintain an open reading frame encoding an integrase-like protein
- PMID: 11058106
- PMCID: PMC113129
- DOI: 10.1093/nar/28.21.4105
A family of developmentally excised DNA elements in Tetrahymena is under selective pressure to maintain an open reading frame encoding an integrase-like protein
Abstract
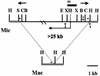
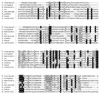
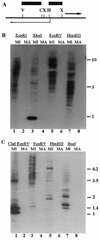
Tlr1 is a member of a family of approximately 20-30 DNA elements that undergo developmentally regulated excision during formation of the macronucleus in the ciliated protozoan TETRAHYMENA: Analysis of sequence internal to the right boundary of Tlr1 revealed the presence of a 2 kb open reading frame (ORF) encoding a deduced protein with similarity to retrotransposon integrases. The ORFs of five unique clones were sequenced. The ORFs have 98% sequence conservation and align without frameshifts, although one has an additional trinucleotide at codon 561. Nucleotide changes among the five clones are highly non-random with respect to the position in the codon and 93% of the nucleotide changes among the five clones encode identical or similar amino acids, suggesting that the ORF has evolved under selective pressure to preserve a functional protein. Nineteen T/C transitions in T/CAA and T/CAG codons suggest selection has occurred in the context of the TETRAHYMENA: genome, where TAA and TAG encode Gln. Similarities between the ORF and those encoding retrotransposon integrases suggest that the Tlr family of elements may encode a polynucleotide transferase. Possible roles for the protein in transposition of the elements within the micronuclear genome and/or their developmentally regulated excision from the macronucleus are discussed.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

