Secondary structure prediction and in vitro accessibility of mRNA as tools in the selection of target sites for ribozymes
- PMID: 11058107
- PMCID: PMC113158
- DOI: 10.1093/nar/28.21.4113
Secondary structure prediction and in vitro accessibility of mRNA as tools in the selection of target sites for ribozymes
Abstract
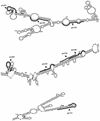
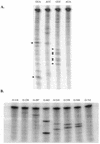
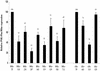
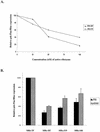
We have investigated the relative merits of two commonly used methods for target site selection for ribozymes: secondary structure prediction (MFold program) and in vitro accessibility assays. A total of eight methylated ribozymes with DNA arms were synthesized and analyzed in a transient co-transfection assay in HeLa cells. Residual expression levels ranging from 23 to 72% were obtained with anti-PSKH1 ribozymes compared to cells transfected with an irrelevant control ribozyme. Ribozyme efficacy depended on both ribozyme concentration and the steady state expression levels of the target mRNA. Allylated ribozymes against a subset of the target sites generally displayed poorer efficacy than their methylated counterparts. This effect appeared to be influenced by in vivo accessibility of the target site. Ribozymes designed on the basis of either selection method displayed a wide range of efficacies with no significant differences in the average activities of the two groups of ribozymes. While in vitro accessibility assays had limited predictive power, there was a significant correlation between certain features of the predicted secondary structure of the target sequence and the efficacy of the corresponding ribozyme. Specifically, ribozyme efficacy appeared to be positively correlated with the presence of short stem regions and helices of low stability within their target sequences. There were no correlations with predicted free energy or loop length.
Figures

Similar articles
-
HIV-1 LTR as a target for synthetic ribozyme-mediated inhibition of gene expression: site selection and inhibition in cell culture.Nucleic Acids Res. 2000 Nov 1;28(21):4059-67. doi: 10.1093/nar/28.21.4059. Nucleic Acids Res. 2000. PMID: 11058100 Free PMC article.
-
Ribozyme expression systems.Methods Mol Biol. 2004;252:195-207. doi: 10.1385/1-59259-746-7:195. Methods Mol Biol. 2004. PMID: 15017050
-
Inhibition of luciferase expression by synthetic hammerhead ribozymes and their cellular uptake.Nucleic Acids Res. 1999 Aug 1;27(15):3159-67. doi: 10.1093/nar/27.15.3159. Nucleic Acids Res. 1999. PMID: 10454613 Free PMC article.
-
Mechanism of action of hammerhead ribozymes and their applications in vivo: rapid identification of functional genes in the post-genome era by novel hybrid ribozyme libraries.Biochem Soc Trans. 2002 Nov;30(Pt 6):1145-9. doi: 10.1042/bst0301145. Biochem Soc Trans. 2002. PMID: 12440992 Review.
-
Ribozymes in the age of molecular therapeutics.Curr Mol Med. 2004 Aug;4(5):489-506. doi: 10.2174/1566524043360410. Curr Mol Med. 2004. PMID: 15267221 Review.
Cited by
-
Profiled support vector machines for antisense oligonucleotide efficacy prediction.BMC Bioinformatics. 2004 Sep 22;5:135. doi: 10.1186/1471-2105-5-135. BMC Bioinformatics. 2004. PMID: 15383156 Free PMC article.
-
Identifying ribozyme-accessible sites using NUH triplet-targeting gapmers.Nucleic Acids Res. 2001 May 1;29(9):1906-14. doi: 10.1093/nar/29.9.1906. Nucleic Acids Res. 2001. PMID: 11328874 Free PMC article.
-
A cellular high-throughput screening approach for therapeutic trans-cleaving ribozymes and RNAi against arbitrary mRNA disease targets.Exp Eye Res. 2016 Oct;151:236-55. doi: 10.1016/j.exer.2016.05.020. Epub 2016 May 25. Exp Eye Res. 2016. PMID: 27233447 Free PMC article.
-
Gene silencing of HIV chemokine receptors using ribozymes and single-stranded antisense RNA.Biochem J. 2006 Mar 1;394(Pt 2):511-8. doi: 10.1042/BJ20051268. Biochem J. 2006. PMID: 16293105 Free PMC article.
-
Ribozyme-based gene-inactivation systems require a fine comprehension of their substrate specificities; the case of delta ribozyme.Curr Med Chem. 2003 Dec;10(23):2589-97. doi: 10.2174/0929867033456486. Curr Med Chem. 2003. PMID: 14529473 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

