Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro
- PMID: 11058110
- PMCID: PMC113145
- DOI: 10.1093/nar/28.21.4138
Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro
Abstract
Error-free lesion bypass and error-prone lesion bypass are important cellular responses to DNA damage during replication, both of which require a DNA polymerase (Pol). To identify lesion bypass DNA polymerases, we have purified human Polkappa encoded by the DINB1 gene and examined its response to damaged DNA templates. Here, we show that human Polkappa is a novel lesion bypass polymerase in vitro. Purified human Polkappa efficiently bypassed a template 8-oxoguanine, incorporating mainly A and less frequently C opposite the lesion. Human Polkappa most frequently incorporated A opposite a template abasic site. Efficient further extension required T as the next template base, and was mediated mainly by a one-nucleotide deletion mechanism. Human Polkappa was able to bypass an acetylaminofluorene-modified G in DNA, incorporating either C or T, and less efficiently A opposite the lesion. Furthermore, human Polkappa effectively bypassed a template (-)-trans-anti-benzo[a]pyrene-N:(2)-dG lesion in an error-free manner by incorporating a C opposite the bulky adduct. In contrast, human Polkappa was unable to bypass a template TT dimer or a TT (6-4) photoproduct, two of the major UV lesions. These results suggest that Polkappa plays an important role in both error-free and error-prone lesion bypass in humans.
Figures
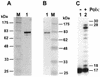
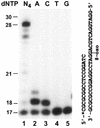
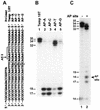
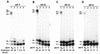



Similar articles
-
Activities of human DNA polymerase kappa in response to the major benzo[a]pyrene DNA adduct: error-free lesion bypass and extension synthesis from opposite the lesion.DNA Repair (Amst). 2002 Jul 17;1(7):559-69. doi: 10.1016/s1568-7864(02)00055-1. DNA Repair (Amst). 2002. PMID: 12509229
-
Translesion synthesis by yeast DNA polymerase zeta from templates containing lesions of ultraviolet radiation and acetylaminofluorene.Nucleic Acids Res. 2001 Jul 1;29(13):2875-83. doi: 10.1093/nar/29.13.2875. Nucleic Acids Res. 2001. PMID: 11433034 Free PMC article.
-
Extending the understanding of mutagenicity: structural insights into primer-extension past a benzo[a]pyrene diol epoxide-DNA adduct.J Mol Biol. 2003 Apr 4;327(4):797-818. doi: 10.1016/s0022-2836(03)00187-6. J Mol Biol. 2003. PMID: 12654264
-
Lesion processing: high-fidelity versus lesion-bypass DNA polymerases.Trends Biochem Sci. 2008 May;33(5):209-19. doi: 10.1016/j.tibs.2008.02.004. Epub 2008 Apr 11. Trends Biochem Sci. 2008. PMID: 18407502 Free PMC article. Review.
-
poliota-dependent lesion bypass in vitro.Mutat Res. 2002 Dec 29;510(1-2):9-22. doi: 10.1016/s0027-5107(02)00248-8. Mutat Res. 2002. PMID: 12459439 Review.
Cited by
-
Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity.Nucleic Acids Res. 2000 Nov 1;28(21):4147-56. doi: 10.1093/nar/28.21.4147. Nucleic Acids Res. 2000. PMID: 11058111 Free PMC article.
-
DNA Damage Tolerance Pathways in Human Cells: A Potential Therapeutic Target.Front Oncol. 2022 Feb 7;11:822500. doi: 10.3389/fonc.2021.822500. eCollection 2021. Front Oncol. 2022. PMID: 35198436 Free PMC article. Review.
-
Generation, repair and replication of guanine oxidation products.Genes Environ. 2017 Aug 1;39:21. doi: 10.1186/s41021-017-0081-0. eCollection 2017. Genes Environ. 2017. PMID: 28781714 Free PMC article. Review.
-
Synthesis and thermodynamic studies of oligodeoxyribonucleotides containing tandem lesions of thymidine glycol and 8-oxo-2'-deoxyguanosine.Chem Res Toxicol. 2006 Jun;19(6):837-43. doi: 10.1021/tx060032l. Chem Res Toxicol. 2006. PMID: 16780363 Free PMC article.
-
Amino acid architecture that influences dNTP insertion efficiency in Y-family DNA polymerase V of E. coli.J Mol Biol. 2009 Sep 18;392(2):270-82. doi: 10.1016/j.jmb.2009.07.016. Epub 2009 Jul 14. J Mol Biol. 2009. PMID: 19607844 Free PMC article.
References
-
- Walker G.C. (1995) Trends Biochem. Sci., 20, 416–420. - PubMed
-
- Goodman M.F. and Tippin,B. (2000) Curr. Opin. Genet. Dev., 10, 162–168. - PubMed
-
- Reuven N.B., Arad,G., Maor-Shoshani,A. and Livneh,Z. (1999) J. Biol. Chem., 274, 31763–31766. - PubMed
-
- Wagner J., Gruz,P., Kim,S.R., Yamada,M., Matsui,K., Fuchs,R.P. and Nohmi,T. (1999) Mol. Cell, 4, 281–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

