Blocking transcription of the human rhodopsin gene by triplex-mediated DNA photocrosslinking
- PMID: 11058128
- PMCID: PMC113126
- DOI: 10.1093/nar/28.21.4283
Blocking transcription of the human rhodopsin gene by triplex-mediated DNA photocrosslinking
Abstract
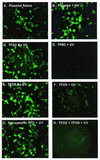
To explore the ability of triplex-forming oligodeoxyribonucleotides (TFOs) to inhibit genes responsible for dominant genetic disorders, we used two TFOs to block expression of the human rhodopsin gene, which encodes a G protein-coupled receptor involved in the blinding disorder autosomal dominant retinitis pigmentosa. Psoralen-modified TFOs and UVA irradiation were used to form photoadducts at two target sites in a plasmid expressing a rhodopsin-EGFP fusion, which was then transfected into HT1080 cells. Each TFO reduced rhodopsin-GFP expression by 70-80%, whereas treatment with both reduced expression by 90%. Expression levels of control genes on either the same plasmid or one co-transfected were not affected by the treatment. Mutations at one TFO target eliminated its effect on transcription, without diminishing inhibition by the other TFO. Northern blots indicated that TFO-directed psoralen photoadducts blocked progression of RNA polymerase, resulting in truncated transcripts. Inhibition of gene expression was not relieved over a 72 h period, suggesting that TFO-induced psoralen lesions are not repaired on this time scale. Irradiation of cells after transfection with plasmid and psoralen-TFOs produced photoadducts inside the cells and also inhibited expression of rhodopsin-EGFP. We conclude that directing DNA damage with psoralen-TFOs is an efficient and specific means for blocking transcription from the human rhodopsin gene.
Figures





Similar articles
-
The nature of dominant mutations of rhodopsin and implications for gene therapy.Mol Neurobiol. 2003 Oct;28(2):149-58. doi: 10.1385/MN:28:2:149. Mol Neurobiol. 2003. PMID: 14576453 Review.
-
[Triple helix: a new promise for gene therapy].Orv Hetil. 2003 Apr 20;144(16):757-63. Orv Hetil. 2003. PMID: 12778626 Hungarian.
-
Psoralen photo-cross-linking by triplex-forming oligonucleotides at multiple sites in the human rhodopsin gene.Biochemistry. 1999 Sep 28;38(39):12850-9. doi: 10.1021/bi9902743. Biochemistry. 1999. PMID: 10504255
-
Psoralen adducts induced by triplex-forming oligonucleotides are refractory to repair in HeLa cells.J Mol Biol. 2000 Feb 18;296(2):373-83. doi: 10.1006/jmbi.1999.3466. J Mol Biol. 2000. PMID: 10669595
-
Gene therapy in animal models of autosomal dominant retinitis pigmentosa.Mol Vis. 2012;18:2479-96. Epub 2012 Oct 6. Mol Vis. 2012. PMID: 23077406 Free PMC article. Review.
Cited by
-
Manipulating the mammalian genome by homologous recombination.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8403-10. doi: 10.1073/pnas.111009698. Proc Natl Acad Sci U S A. 2001. PMID: 11459982 Free PMC article. Review.
-
Knock-in human rhodopsin-GFP fusions as mouse models for human disease and targets for gene therapy.Proc Natl Acad Sci U S A. 2004 Jun 15;101(24):9109-14. doi: 10.1073/pnas.0403149101. Epub 2004 Jun 7. Proc Natl Acad Sci U S A. 2004. PMID: 15184660 Free PMC article.
-
Sequence-specific triple helix formation with genomic DNA.Biochemistry. 2007 Oct 9;46(40):11240-52. doi: 10.1021/bi700580y. Epub 2007 Sep 11. Biochemistry. 2007. PMID: 17845009 Free PMC article.
-
The nature of dominant mutations of rhodopsin and implications for gene therapy.Mol Neurobiol. 2003 Oct;28(2):149-58. doi: 10.1385/MN:28:2:149. Mol Neurobiol. 2003. PMID: 14576453 Review.
-
Non-canonical DNA structures: Diversity and disease association.Front Genet. 2022 Sep 5;13:959258. doi: 10.3389/fgene.2022.959258. eCollection 2022. Front Genet. 2022. PMID: 36134025 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

