Characterization of the interaction between alphaCP(2) and the 3'-untranslated region of collagen alpha1(I) mRNA
- PMID: 11058131
- PMCID: PMC113122
- DOI: 10.1093/nar/28.21.4306
Characterization of the interaction between alphaCP(2) and the 3'-untranslated region of collagen alpha1(I) mRNA
Abstract
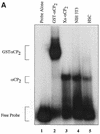
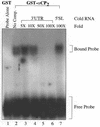
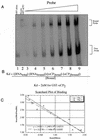
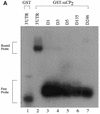
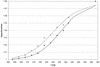
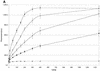
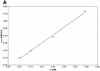
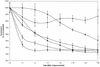
Activated hepatic stellate cells produce increased type I collagen in hepatic fibrosis. The increase in type I collagen protein results from an increase in mRNA levels that is mainly mediated by increased mRNA stability. Protein-RNA interactions in the 3'-UTR of the collagen alpha1(I) mRNA correlate with stabilization of the mRNA during hepatic stellate cell activation. A component of the binding complex is alphaCP(2). Recombinant alphaCP(2) is sufficient for binding to the 3'-UTR of collagen alpha1(I). To characterize the binding affinity of and specificity for alphaCP(2), we performed electrophoretic mobility shift assays using the poly(C)-rich sequence in the 3'-UTR of collagen alpha1(I) as probe. The binding affinity of alphaCP(2) for the 3'-UTR sequence is approximately 2 nM in vitro and the wild-type 3' sequence binds with high specificity. Furthermore, we demonstrate a system for detecting protein-nucleotide interactions that is suitable for high throughput assays using molecular beacons. Molecular beacons, developed for DNA-DNA hybridization, are oligonucleotides with a fluorophore and quencher brought together by a hairpin sequence. Fluorescence increases when the hairpin is disrupted by binding to an antisense sequence or interaction with a protein. Molecular beacons displayed a similar high affinity for binding to recombinant alphaCP(2) to the wild-type 3' sequence, although the kinetics of binding were slower.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

