tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding
- PMID: 11060012
- PMCID: PMC305789
- DOI: 10.1093/emboj/19.21.5599
tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding
Abstract
The 2.2 A crystal structure of a ternary complex formed by yeast arginyl-tRNA synthetase and its cognate tRNA(Arg) in the presence of the L-arginine substrate highlights new atomic features used for specific substrate recognition. This first example of an active complex formed by a class Ia aminoacyl-tRNA synthetase and its natural cognate tRNA illustrates additional strategies used for specific tRNA selection. The enzyme specifically recognizes the D-loop and the anticodon of the tRNA, and the mutually induced fit produces a conformation of the anticodon loop never seen before. Moreover, the anticodon binding triggers conformational changes in the catalytic center of the protein. The comparison with the 2.9 A structure of a binary complex formed by yeast arginyl-tRNA synthetase and tRNA(Arg) reveals that L-arginine binding controls the correct positioning of the CCA end of the tRNA(Arg). Important structural changes induced by substrate binding are observed in the enzyme. Several key residues of the active site play multiple roles in the catalytic pathway and thus highlight the structural dynamics of the aminoacylation reaction.
Figures
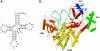
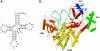







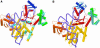
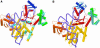
References
-
- Auffinger P. and Westhof,E. (1998) Hydration of RNA base pairs. J. Biomol. Struct. Dyn., 16, 693–707. - PubMed
-
- Belrhali H., Yaremchuck,A., Tukalo,M., Berthet-Colominas,C., Rasmussen,B., Bosecke,P., Diat,O. and Cusack,S. (1995) The structural basis for seryl-adenylate and Ap4A synthesis by seryl-tRNA synthetase. Structure, 3, 341–352. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

