A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae
- PMID: 11060033
- PMCID: PMC305808
- DOI: 10.1093/emboj/19.21.5824
A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae
Abstract
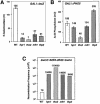
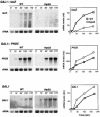
Transcription-induced recombination has been reported in all organisms from bacteria to mammals. We have shown previously that the yeast genes HPR1 and THO2 may be keys to the understanding of transcription-associated recombination, as they both affect transcription elongation and hyper-recombination in a concerted manner. Using a yeast strain that has the wild-type THO2 gene replaced by one encoding a His(6)-HA-tagged version, we have isolated an oligomeric complex containing four proteins: Tho2, Hpr1, Mft1 and a novel protein that we have named Thp2. We have reciprocally identified a complex containing Hpr1, Tho2 and Mft1 using anti-Mft1 antibodies in immunoprecipitation experiments. The protein complex is mainly nuclear; therefore, Tho2 and Hpr1 are physically associated. Like hpr1Delta and tho2Delta cells, mft1Delta and thp2Delta cells show mitotic hyper- recombination and impaired transcription elongation, in particular, through the bacterial lacZ sequence. Hyper-recombination conferred by mft1Delta and thp2Delta is only observed in DNA regions under transcription conditions. We propose that this protein complex acts as a functional unit connecting transcription elongation with the incidence of mitotic recombination.
Figures
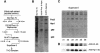





References
-
- Beilharz T., Beddoe,T., Landl,K., Cartwright,P. and Lithgow,T. (1997) The protein encoded by the MFT1 gene is a targeting factor for mitochondrial precursor proteins and not a core ribosomal protein. FEBS Lett., 407, 220–224. - PubMed
-
- Blackwell T.K., Moore,M.W., Yancopoulos,G.D., Suh,H., Lutzker,S., Selsing,E. and Alt,F.W. (1986) Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature, 324, 585–589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

