Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors
- PMID: 11060040
- PMCID: PMC305781
- DOI: 10.1093/emboj/19.21.5895
Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors
Abstract
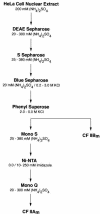
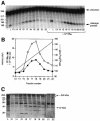
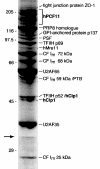
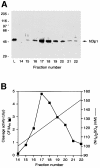
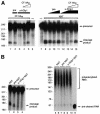
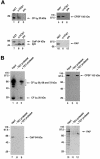
Six different protein factors are required in vitro for 3' end formation of mammalian pre-mRNAs by endonucleolytic cleavage and polyadenylation. Five of the factors have been purified and most of their components cloned, but cleavage factor II(m) (CF II(m)) remained uncharacterized. We have purified CF II(m) from HeLa cell nuclear extract by several chromatographic steps. During purification, CF II(m) activity separated into two components, one essential (CF IIA(m)) and one stimulatory (CF IIB(m)) for the cleavage reaction. CF IIA(m) fractions contain the human homologs of two yeast 3' end processing factors, Pcf11p and Clp1p, as well as cleavage factor I(m) (CF I(m)) and several splicing and transcription factors. We report the cloning of hClp1 and show that it is a genuine subunit of CF IIA(m). Antibodies directed against hClp1 deplete cleavage activity, but not polyadenylation activity from HeLa cell nuclear extract. hClp1 interacts with CF I(m) and the cleavage and polyadenylation specificity factor CPSF, suggesting that it bridges these two 3' end processing factors within the cleavage complex.
Figures

References
-
- Barabino S.M. and Keller,W. (1999) Last but not least: regulated poly(A) tail formation. Cell, 99, 9–11. - PubMed
-
- Barabino S.M., Hübner,W., Jenny,A., Minvielle-Sebastia,L. and Keller,W. (1997) The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev., 11, 1703–1716. - PubMed
-
- Bienroth S., Wahle,E., Suter-Crazzolara,C. and Keller,W. (1991) Purification of the cleavage and polyadenylation factor involved in the 3′-end processing of messenger RNA precursors. J. Biol. Chem., 266, 19768–19776. - PubMed
-
- Fountoulakis M. and Langen,H. (1997) Identification of proteins by matrix-assisted laser desorption ionization-mass spectrometry following in-gel digestion in low-salt, nonvolatile buffer and simplified peptide recovery. Anal. Biochem., 250, 153–156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

