Identification of ciprofloxacin-resistant Campylobacter jejuni by use of a fluorogenic PCR assay
- PMID: 11060054
- PMCID: PMC87527
- DOI: 10.1128/JCM.38.11.3971-3978.2000
Identification of ciprofloxacin-resistant Campylobacter jejuni by use of a fluorogenic PCR assay
Abstract
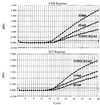
Fluoroquinolones are one class of antimicrobial agents commonly used to treat severe Campylobacter jejuni infection. C. jejuni strains resistant to high levels of the fluoroquinolone ciprofloxacin (MIC >/=16 microg/ml) have been predominantly characterized with a C-->T transition in codon 86 of gyrA. The gyrA gene encodes one subunit of DNA gyrase, which is a primary target for fluoroquinolone antibiotics. This study establishes a rapid PCR-based TaqMan method for identifying ciprofloxacin-resistant C. jejuni strains that carry the C-->T transition in codon 86 of gyrA. The assay uses real-time detection, eliminating the need for gel electrophoresis. Optimization of the assay parameters using purified Campylobacter DNA resulted in the ability to detect femtogram levels of DNA. The method should be useful for monitoring the development of ciprofloxacin resistance in C. jejuni. Compiled nucleotide sequence data on the quinolone resistance-determining region of gyrA in Campylobacter indicate that sequence comparison of this region is a useful method for tentative identification of Campylobacter isolates at the species level.
Figures



Similar articles
-
Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis.J Clin Microbiol. 1999 Oct;37(10):3276-80. doi: 10.1128/JCM.37.10.3276-3280.1999. J Clin Microbiol. 1999. PMID: 10488192 Free PMC article.
-
Detection of ciprofloxacin resistance mutations in Campylobacter jejuni gyrA by nonradioisotopic single-strand conformation polymorphism and direct DNA sequencing.J Clin Lab Anal. 1996;10(3):129-33. doi: 10.1002/(SICI)1098-2825(1996)10:3<129::AID-JCLA3>3.0.CO;2-6. J Clin Lab Anal. 1996. PMID: 8731499
-
Detection of gyrA mutation among clinical isolates of Campylobacter jejuni isolated in Egypt by MAMA-PCR.J Infect Dev Ctries. 2010 Oct 4;4(9):546-54. doi: 10.3855/jidc.963. J Infect Dev Ctries. 2010. PMID: 21045366
-
High level of ciprofloxacin resistance and its molecular background among Campylobacter jejuni strains isolated in the United Arab Emirates.J Med Microbiol. 2006 Nov;55(Pt 11):1533-1538. doi: 10.1099/jmm.0.46744-0. J Med Microbiol. 2006. PMID: 17030913
-
Mechanisms of antibiotic resistance in Campylobacter species.Antimicrob Agents Chemother. 1988 Aug;32(8):1107-12. doi: 10.1128/AAC.32.8.1107. Antimicrob Agents Chemother. 1988. PMID: 3056250 Free PMC article. Review. No abstract available.
Cited by
-
Incidence and mechanism of ciprofloxacin resistance in Campylobacter spp. isolated from commercial poultry flocks in the United Kingdom before, during, and after fluoroquinolone treatment.Antimicrob Agents Chemother. 2005 Feb;49(2):699-707. doi: 10.1128/AAC.49.2.699-707.2005. Antimicrob Agents Chemother. 2005. PMID: 15673754 Free PMC article.
-
Contrasting immune responses mediate Campylobacter jejuni-induced colitis and autoimmunity.Mucosal Immunol. 2014 Jul;7(4):802-17. doi: 10.1038/mi.2013.97. Epub 2013 Nov 13. Mucosal Immunol. 2014. PMID: 24220299 Free PMC article.
-
Massive microbiological groundwater contamination associated with a waterborne outbreak in Lake Erie, South Bass Island, Ohio.Environ Health Perspect. 2007 Jun;115(6):856-64. doi: 10.1289/ehp.9430. Epub 2007 Feb 6. Environ Health Perspect. 2007. PMID: 17589591 Free PMC article.
-
PCR-restriction fragment length polymorphism assay for detection of gyrA mutations associated with fluoroquinolone resistance in Campylobacter coli.Antimicrob Agents Chemother. 2004 Dec;48(12):4886-8. doi: 10.1128/AAC.48.12.4886-4888.2004. Antimicrob Agents Chemother. 2004. PMID: 15561873 Free PMC article.
-
Concurrent quantitation of total campylobacter and total ciprofloxacin-resistant campylobacter loads in rinses from retail raw chicken carcasses from 2001 to 2003 by direct plating at 42 degrees C.Appl Environ Microbiol. 2005 Aug;71(8):4510-5. doi: 10.1128/AEM.71.8.4510-4515.2005. Appl Environ Microbiol. 2005. PMID: 16085843 Free PMC article.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997.
-
- Belgrader P, Benett W, Hadley D, Richards J, Stratton P, Mariella R, Jr, Milanovich F. PCR detection of bacteria in seven minutes. Science. 1999;284:449–450. - PubMed
-
- Blaser M J. Epidemiologic and clinical features of Campylobacter jejuni infections. J Infect Dis. 1997;176(Suppl. 2):S103–S105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

