ACAPs are arf6 GTPase-activating proteins that function in the cell periphery
- PMID: 11062263
- PMCID: PMC2185579
- DOI: 10.1083/jcb.151.3.627
ACAPs are arf6 GTPase-activating proteins that function in the cell periphery
Abstract
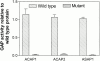
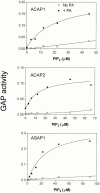
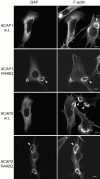
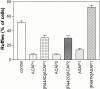
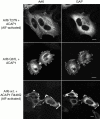
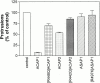
The GTP-binding protein ADP-ribosylation factor 6 (Arf6) regulates endosomal membrane trafficking and the actin cytoskeleton in the cell periphery. GTPase-activating proteins (GAPs) are critical regulators of Arf function, controlling the return of Arf to the inactive GDP-bound state. Here, we report the identification and characterization of two Arf6 GAPs, ACAP1 and ACAP2. Together with two previously described Arf GAPs, ASAP1 and PAP, they can be grouped into a protein family defined by several common structural motifs including coiled coil, pleckstrin homology, Arf GAP, and three complete ankyrin-repeat domains. All contain phosphoinositide-dependent GAP activity. ACAP1 and ACAP2 are widely expressed and occur together in the various cultured cell lines we examined. Similar to ASAP1, ACAP1 and ACAP2 were recruited to and, when overexpressed, inhibited the formation of platelet-derived growth factor (PDGF)-induced dorsal membrane ruffles in NIH 3T3 fibroblasts. However, in contrast with ASAP1, ACAP1 and ACAP2 functioned as Arf6 GAPs. In vitro, ACAP1 and ACAP2 preferred Arf6 as a substrate, rather than Arf1 and Arf5, more so than did ASAP1. In HeLa cells, overexpression of either ACAP blocked the formation of Arf6-dependent protrusions. In addition, ACAP1 and ACAP2 were recruited to peripheral, tubular membranes, where activation of Arf6 occurs to allow membrane recycling back to the plasma membrane. ASAP1 did not inhibit Arf6-dependent protrusions and was not recruited by Arf6 to tubular membranes. The additional effects of ASAP1 on PDGF-induced ruffling in fibroblasts suggest that multiple Arf GAPs function coordinately in the cell periphery.
Figures





Similar articles
-
Proteomic identification and functional characterization of a novel ARF6 GTPase-activating protein, ACAP4.Mol Cell Proteomics. 2006 Aug;5(8):1437-49. doi: 10.1074/mcp.M600050-MCP200. Epub 2006 May 30. Mol Cell Proteomics. 2006. PMID: 16737952
-
AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton.J Biol Chem. 2002 Dec 13;277(50):48965-75. doi: 10.1074/jbc.M202969200. Epub 2002 Oct 17. J Biol Chem. 2002. PMID: 12388557
-
DEF-1/ASAP1 is a GTPase-activating protein (GAP) for ARF1 that enhances cell motility through a GAP-dependent mechanism.J Biol Chem. 2002 Mar 8;277(10):7962-9. doi: 10.1074/jbc.M109149200. Epub 2001 Dec 31. J Biol Chem. 2002. PMID: 11773070
-
Arf GAPs as Regulators of the Actin Cytoskeleton-An Update.Int J Mol Sci. 2019 Jan 21;20(2):442. doi: 10.3390/ijms20020442. Int J Mol Sci. 2019. PMID: 30669557 Free PMC article. Review.
-
Arf GAPs: A family of proteins with disparate functions that converge on a common structure, the integrin adhesion complex.Small GTPases. 2019 Jul;10(4):280-288. doi: 10.1080/21541248.2017.1299271. Epub 2017 Mar 31. Small GTPases. 2019. PMID: 28362242 Free PMC article. Review.
Cited by
-
Allosteric properties of PH domains in Arf regulatory proteins.Cell Logist. 2016 Apr 26;6(2):e1181700. doi: 10.1080/21592799.2016.1181700. eCollection 2016 Apr-Jun. Cell Logist. 2016. PMID: 27294009 Free PMC article. Review.
-
Identification of an intramolecular interaction important for the regulation of GIT1 functions.Mol Biol Cell. 2007 Dec;18(12):5124-38. doi: 10.1091/mbc.e07-06-0550. Epub 2007 Sep 26. Mol Biol Cell. 2007. PMID: 17898078 Free PMC article.
-
Arf1 and Arf6 promote ventral actin structures formed by acute activation of protein kinase C and Src.Cytoskeleton (Hoboken). 2014 Jun;71(6):380-94. doi: 10.1002/cm.21181. Epub 2014 Jun 26. Cytoskeleton (Hoboken). 2014. PMID: 24916416 Free PMC article.
-
Parkin-dependent ubiquitination of TAX1BP1 directs efficient autophagic removal of defective mitochondria.bioRxiv [Preprint]. 2025 May 19:2025.05.16.654474. doi: 10.1101/2025.05.16.654474. bioRxiv. 2025. PMID: 40475537 Free PMC article. Preprint.
-
A class I ADP-ribosylation factor GTPase-activating protein is critical for maintaining directional root hair growth in Arabidopsis.Plant Physiol. 2008 Aug;147(4):1659-74. doi: 10.1104/pp.108.119529. Epub 2008 Jun 6. Plant Physiol. 2008. PMID: 18539780 Free PMC article.
References
-
- Bagrodia S., Bailey D., Lenard Z., Hart M., Guan J.L., Premont R.T., Taylor S.J., Cerione R.A. A tyrosine-phosphorylated protein that binds to an important regulatory region on the cool family of p21-activated kinase-binding proteins. J. Biol. Chem. 1999;274:22393–22400. - PubMed
-
- Balch W.E., Kahn R.A., Schwaninger R. ADP-ribosylation factor is required for vesicular trafficking between the endoplasmic reticulum and the cis-Golgi compartment. J. Biol. Chem. 1992;267:13053–13061. - PubMed
-
- Billuart P., Bienvenu T., Ronce N., des Portes V., Vinet M.C., Zemni R., Crollius H.R., Carrie A., Fauchereau F., Cherry M., Briault S., Hamel B., Fryns J.P., Beldjord C., Kahn A., Moraine C., Chelly J. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392:923–926. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

