A GNAS1 imprinting defect in pseudohypoparathyroidism type IB
- PMID: 11067869
- PMCID: PMC301417
- DOI: 10.1172/JCI10431
A GNAS1 imprinting defect in pseudohypoparathyroidism type IB
Abstract
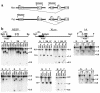
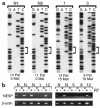
Pseudohypoparathyroidism type IB (PHPIB) is characterized by renal resistance to parathyroid hormone (PTH) and the absence of other endocrine or physical abnormalities. Familial PHPIB has been mapped to 20q13, near GNAS1, which encodes G(s)alpha, the G protein alpha-subunit required for receptor-stimulated cAMP generation. However, G(s)alpha function is normal in blood cells from PHPIB patients, ruling out mutations within the G(s)alpha coding region. In mice G(s)alpha is expressed only from the maternal allele in renal proximal tubules (the site of PTH action) but is biallelically expressed in most other tissues. Studies in patients with Albright hereditary osteodystrophy suggest a similar G(s)alpha imprinting pattern in humans. Here we identify a region upstream of the G(s)alpha promoter that is normally methylated on the maternal allele and unmethylated on the paternal allele, but that is unmethylated on both alleles in all 13 PHPIB patients studied. Within this region is an alternative promoter and first exon (exon 1A), generating transcripts that are normally expressed only from the paternal allele, but that are biallelically expressed in PHPIB patients. Therefore, PHPIB is associated with a paternal-specific imprinting pattern of the exon 1A region on both alleles, which may lead to decreased G(s)alpha expression in renal proximal tubules. We propose that loss of exon 1A imprinting is the cause of PHPIB.
Figures


Comment in
-
Imprints of disease at GNAS1.J Clin Invest. 2001 Apr;107(7):793-4. doi: 10.1172/JCI12645. J Clin Invest. 2001. PMID: 11285295 Free PMC article. Review. No abstract available.
References
-
- Levine MA, et al. Resistance to multiple hormones in patients with pseudohypoparathyroidism. Association with deficient activity of guanine nucleotide regulatory protein. Am J Med. 1983;74:545–556. - PubMed
-
- Schipani E, et al. Pseudohypoparathyroidism type Ib is not caused by mutations in the coding exons of the human parathyroid hormone (PTH)/PTH-related peptide receptor gene. J Clin Endocrinol Metab. 1995;80:1611–1621. - PubMed
-
- Weinstein LS, Yu S. The role of genomic imprinting of Gsα in the pathogenesis of Albright hereditary osteodystrophy. Trends Endocrinol Metab. 1999;10:81–85. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

