The bile acid synthetic gene 3beta-hydroxy-Delta(5)-C(27)-steroid oxidoreductase is mutated in progressive intrahepatic cholestasis
- PMID: 11067870
- PMCID: PMC301421
- DOI: 10.1172/JCI10902
The bile acid synthetic gene 3beta-hydroxy-Delta(5)-C(27)-steroid oxidoreductase is mutated in progressive intrahepatic cholestasis
Abstract
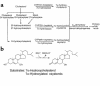
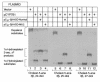
We used expression cloning to isolate cDNAs encoding a microsomal 3beta-hydroxy-Delta(5)-C(27)-steroid oxidoreductase (C(27) 3beta-HSD) that is expressed predominantly in the liver. The predicted product shares 34% sequence identity with the C(19) and C(21) 3beta-HSD enzymes, which participate in steroid hormone metabolism. When transfected into cultured cells, the cloned C(27) 3beta-HSD cDNA encodes an enzyme that is active against four 7alpha-hydroxylated sterols, indicating that a single C(27) 3beta-HSD enzyme can participate in all known pathways of bile acid synthesis. The expressed enzyme did not metabolize several different C(19/21) steroids as substrates. The levels of hepatic C(27) 3beta-HSD mRNA in the mouse are not sexually dimorphic and do not change in response to dietary cholesterol or to changes in bile acid pool size. The corresponding human gene on chromosome 16p11.2-12 contains six exons and spans 3 kb of DNA, and we identified a 2-bp deletion in the C27 3beta-HSD gene of a patient with neonatal progressive intrahepatic cholestasis. This mutation eliminates the activity of the enzyme in transfected cells. These findings establish the central role of C(27) 3beta-HSD in the biosynthesis of bile acids and provide molecular tools for the diagnosis of a third type of neonatal progressive intrahepatic cholestasis associated with impaired bile acid synthesis.
Figures




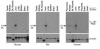


References
-
- Russell DW, Setchell KDR. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. - PubMed
-
- Gourley, G.R. 1994. Bilirubin metabolism and neonatal jaundice. In Liver disease in children. F.J. Suchy, editor. Mosby. St. Louis, Missouri, USA. 105–125.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

