Relative replication fitness of a high-level 3'-azido-3'-deoxythymidine-resistant variant of human immunodeficiency virus type 1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr-->Gly) at codon 69
- PMID: 11069990
- PMCID: PMC113175
- DOI: 10.1128/jvi.74.23.10958-10964.2000
Relative replication fitness of a high-level 3'-azido-3'-deoxythymidine-resistant variant of human immunodeficiency virus type 1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr-->Gly) at codon 69
Abstract
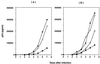
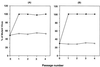
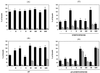
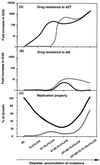
The combination of an amino acid deletion at codon 67 (delta 67) and Thr-to-Gly change at codon 69 (T69G) in the reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1) is associated with high-level resistance to multiple RT inhibitors. To determine the relative contributions of the delta 67 and T69G mutations on viral fitness, we performed a series of studies of HIV replication using recombinant variants. A high-level 3'-azido-3'-deoxythymidine (AZT)-resistant variant containing delta 67 plus T69G/K70R/L74I/K103N/T215F/K219Q in RT replicated as efficiently as wild-type virus (Wt). In contrast, the construct without delta 67 exhibited impaired replication (23% of growth of Wt). A competitive fitness study failed to reveal any differences in replication rates between the delta 67+T69G/K70R/L74I/K103N/T215F/+ ++K219Q mutant and Wt. Evaluation of proviral DNA sequences over a 3-year period in a patient harboring the multiresistant HIV revealed that the T69G mutation emerged in the context of a D67N/K70R/T215F/K219Q mutant backbone prior to appearance of the delta 67 deletion. To assess the impact of this stepwise accumulation of mutations on viral replication, a series of recombinant variants was constructed and analyzed for replication competence. The T69G mutation was found to confer 2',3'-dideoxyinosine resistance at the expense of fitness. Subsequently, the development of the delta 67 deletion led to a virus with improved replication and high-level AZT resistance.
Figures
References
-
- AIDS Clinical Trials Group. Virology manual for HIV laboratories. [Online.] Bethesda, Md: Division of AIDS, National Institute of Allergy and Infectious Diseases; 1997. http://www.niaid.nih.gov./daids/vir manual.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

