The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth
- PMID: 11070009
- PMCID: PMC113196
- DOI: 10.1128/jvi.74.23.11129-11136.2000
The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth
Abstract
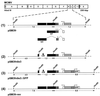
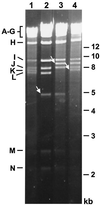
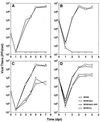
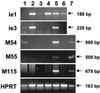
The significance of the major immediate-early gene ie3 of mouse cytomegalovirus (MCMV) and that of the corresponding ie2 gene of human cytomegalovirus to viral replication are not known. To investigate the function of the MCMV IE3 regulatory protein, we generated two different MCMV recombinants that contained a large deletion in the IE3 open reading frame (ORF). The mutant genomes were constructed by the bacterial artificial chromosome mutagenesis technique, and MCMV ie3 deletion mutants were reconstituted on a mouse fibroblast cell line that expresses the MCMV major immediate-early genes. The ie3 deletion mutants failed to replicate on normal mouse fibroblasts even when a high multiplicity of infection was used. The replication defect was rescued when the IE3 protein was provided in trans by a complementing cell line. A revertant virus in which the IE3 ORF was restored was able to replicate with wild-type kinetics in normal mouse fibroblasts, providing evidence that the defective growth phenotype of the ie3 mutants was due to disruption of the ie3 gene. To characterize the point of restriction in viral replication that is controlled by ie3, we analyzed the pattern of expression of selective early (beta) and late (gamma) genes. While we could detect transcripts for the immediate-early gene ie1 in cells infected with the ie3 mutants, we failed to detect transcripts for representative beta and gamma genes. These data demonstrate that the MCMV transactivator IE3 plays an indispensable role during viral replication in tissue culture, implicating a similar role for the human CMV ie2 gene product. To our knowledge, the ie3 deletion mutants represent the first MCMV recombinants isolated that contain a disruption of an essential gene.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

