Repair of gaps in retroviral DNA integration intermediates
- PMID: 11070016
- PMCID: PMC113210
- DOI: 10.1128/jvi.74.23.11191-11200.2000
Repair of gaps in retroviral DNA integration intermediates
Abstract
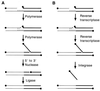
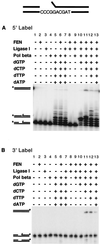
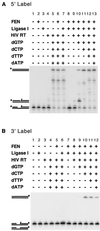
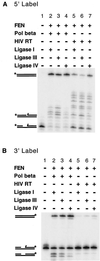
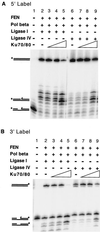
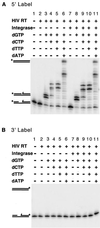
Diverse mobile DNA elements are believed to pirate host cell enzymes to complete DNA transfer. Prominent examples are provided by retroviral cDNA integration and transposon insertion. These reactions initially involve the attachment of each element 3' DNA end to staggered sites in the host DNA by element-encoded integrase or transposase enzymes. Unfolding of such intermediates yields DNA gaps at each junction. It has been widely assumed that host DNA repair enzymes complete attachment of the remaining DNA ends, but the enzymes involved have not been identified for any system. We have synthesized DNA substrates containing the expected gap and 5' two-base flap structure present in retroviral integration intermediates and tested candidate enzymes for the ability to support repair in vitro. We find three required activities, two of which can be satisfied by multiple enzymes. These are a polymerase (polymerase beta, polymerase delta and its cofactor PCNA, or reverse transcriptase), a nuclease (flap endonuclease), and a ligase (ligase I, III, or IV and its cofactor XRCC4). A proposed pathway involving retroviral integrase and reverse transcriptase did not carry out repair under the conditions tested. In addition, prebinding of integrase protein to gapped DNA inhibited repair reactions, indicating that gap repair in vivo may require active disassembly of the integrase complex.
Figures


References
-
- Allen P, Worland S, Gold L. Isolation of high-affinity RNA ligands to HIV-1 integrase from a random pool. Virology. 1995;209:327–336. - PubMed
-
- Baker T A, Kremenstova E, Luo L. Complete transposition requires four active monomers in the Mu transposase tetramer. Genes Dev. 1994;8:2416–2428. - PubMed
-
- Baker T A, Mizuuchi M, Savilahti H, Mizuuchi K. Division of labor among monomers within the Mu transposase tetramer. Cell. 1993;74:723–733. - PubMed
-
- Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

