Open reading frame S/L of varicella-zoster virus encodes a cytoplasmic protein expressed in infected cells
- PMID: 11070031
- PMCID: PMC113236
- DOI: 10.1128/jvi.74.23.11311-11321.2000
Open reading frame S/L of varicella-zoster virus encodes a cytoplasmic protein expressed in infected cells
Abstract
We report the discovery of a novel gene in the varicella-zoster virus (VZV) genome, designated open reading frame (ORF) S/L. This gene, located at the left end of the prototype VZV genome isomer, expresses a polyadenylated mRNA containing a splice within the 3' untranslated region in virus-infected cells. Sequence analysis reveals significant differences between the ORF S/Ls of wild-type and attenuated strains of VZV. Antisera raised to a bacterially expressed portion of ORF S/L reacted specifically with a 21-kDa protein synthesized in cells infected with a VZV clinical isolate and with the original vaccine strain of VZV (Oka-ATCC). Cells infected with other VZV strains, including a wild-type strain that has been extensively passaged in tissue culture and commercially produced vaccine strains of Oka, synthesize a family of proteins ranging in size from 21 to 30 kDa that react with the anti-ORF S/L antiserum. MeWO cells infected with recombinant VZV harboring mutations in the C-terminal region of the ORF S/L gene lost adherence to the stratum and adjacent cells, resulting in an altered plaque morphology. Immunohistochemical analysis of VZV-infected cells demonstrated that ORF S/L protein localizes to the cytoplasm. ORF S/L protein was present in skin lesions of individuals with primary or reactivated infection and in the neurons of a dorsal root ganglion during virus reactivation.
Figures



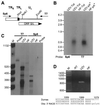

 ) containing a novel
EcoRV site replacing the 120 bp between the EcoRI
and BspEI sites of Oka. Images of the autoradiograms and
stained gels were generated as for Fig. 3.
) containing a novel
EcoRV site replacing the 120 bp between the EcoRI
and BspEI sites of Oka. Images of the autoradiograms and
stained gels were generated as for Fig. 3.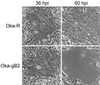




Similar articles
-
Sequence analysis of the leftward end of simian varicella virus (EcoRI-I fragment) reveals the presence of an 8-bp repeat flanking the unique long segment and an 881-bp open-reading frame that is absent in the varicella zoster virus genome.Virology. 2000 Sep 1;274(2):420-8. doi: 10.1006/viro.2000.0465. Virology. 2000. PMID: 10964784
-
The glycoprotein products of varicella-zoster virus gene 14 and their defective accumulation in a vaccine strain (Oka).J Virol. 1990 Sep;64(9):4540-8. doi: 10.1128/JVI.64.9.4540-4548.1990. J Virol. 1990. PMID: 2166829 Free PMC article.
-
New method of differentiating wild-type varicella-zoster virus (VZV) strains from Oka varicella vaccine strain by VZV ORF 6-based PCR and restriction fragment length polymorphism analysis.J Clin Virol. 2004 Feb;29(2):113-9. doi: 10.1016/s1386-6532(03)00112-4. J Clin Virol. 2004. PMID: 14747030
-
Molecular analysis of the Oka vaccine strain of varicella-zoster virus.J Infect Dis. 2008 Mar 1;197 Suppl 2:S45-8. doi: 10.1086/522122. J Infect Dis. 2008. PMID: 18419407 Review.
-
Mutagenesis of the varicella-zoster virus genome: lessons learned.Arch Virol Suppl. 2001;(17):91-7. doi: 10.1007/978-3-7091-6259-0_10. Arch Virol Suppl. 2001. PMID: 11339555 Review.
Cited by
-
Genetic analysis of varicella-zoster virus ORF0 to ORF4 by use of a novel luciferase bacterial artificial chromosome system.J Virol. 2007 Sep;81(17):9024-33. doi: 10.1128/JVI.02666-06. Epub 2007 Jun 20. J Virol. 2007. PMID: 17581997 Free PMC article.
-
Review: The neurobiology of varicella zoster virus infection.Neuropathol Appl Neurobiol. 2011 Aug;37(5):441-63. doi: 10.1111/j.1365-2990.2011.01167.x. Neuropathol Appl Neurobiol. 2011. PMID: 21342215 Free PMC article. Review.
-
Mutational analysis of the varicella-zoster virus ORF62/63 intergenic region.J Virol. 2006 Mar;80(6):3116-21. doi: 10.1128/JVI.80.6.3116-3121.2006. J Virol. 2006. PMID: 16501125 Free PMC article.
-
Varicella-zoster virus T cell tropism and the pathogenesis of skin infection.Curr Top Microbiol Immunol. 2010;342:189-209. doi: 10.1007/82_2010_29. Curr Top Microbiol Immunol. 2010. PMID: 20397071 Free PMC article. Review.
-
Severe Herpes Zoster Following Varicella Vaccination in Immunocompetent Young Children.J Child Neurol. 2019 Mar;34(4):184-188. doi: 10.1177/0883073818821498. Epub 2019 Jan 10. J Child Neurol. 2019. PMID: 30628536 Free PMC article.
References
-
- Centers for Disease Control and Prevention. Varicella-related deaths among adults—United States 1997. Morbid Mortal Wkly Rep. 1997;46:409–412. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

