The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle
- PMID: 11071884
- PMCID: PMC1350968
- DOI: 10.1074/jbc.M004895200
The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle
Abstract
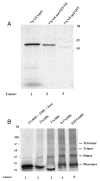
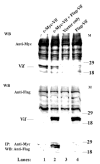
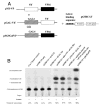
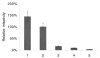
The Vif (virion infectivity factor protein of human immunodeficiency virus type I (HIV-1) is essential for viral replication in vivo and productive infection of peripheral blood mononuclear cells, macrophages, and H9 T-cells. However, the molecular mechanism(s) of Vif remains unknown and needs to be further determined. In this report, we show that, like many other proteins encoded by HIV-1, Vif proteins possess a strong tendency toward self-association. In relatively native conditions, Vif proteins formed multimers in vitro, including dimers, trimers, or tetramers. Through in vivo binding assays such as coimmunoprecipitation and the mammalian two-hybrid system, we also demonstrated that Vif proteins could interact with each other within a cell, indicating that the multimerization of Vif proteins is not simply due to fortuitous aggregation. Further studies indicated that the domain affecting Vif self-association is located at the C terminus of this protein, especially the proline-enriched 151-164 region. Moreover, we found that a Vif mutant with deletion at amino acid 151-164 was unable to rescue the infectivity of vif-defective viruses generated from H9 T-cells, suggesting that the multimerization of Vif proteins could be important for Vif function in the viral life cycle. Our studies identified a new feature of Vif and should accelerate our understanding of its role in HIV-1 pathogenesis.
Figures

Similar articles
-
Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins.J Biol Chem. 2003 Feb 21;278(8):6596-602. doi: 10.1074/jbc.M210164200. Epub 2002 Dec 11. J Biol Chem. 2003. PMID: 12480936 Free PMC article.
-
Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process.J Virol. 2000 Sep;74(18):8252-61. doi: 10.1128/jvi.74.18.8252-8261.2000. J Virol. 2000. PMID: 10954522 Free PMC article.
-
Ring finger protein ZIN interacts with human immunodeficiency virus type 1 Vif.J Virol. 2004 Oct;78(19):10574-81. doi: 10.1128/JVI.78.19.10574-10581.2004. J Virol. 2004. PMID: 15367624 Free PMC article.
-
The role of Vif during HIV-1 infection: interaction with novel host cellular factors.J Clin Virol. 2003 Feb;26(2):143-52. doi: 10.1016/s1386-6532(02)00113-0. J Clin Virol. 2003. PMID: 12600646 Review.
-
Cell-dependent function of HIV-1 Vif for virus replication (Review).Int J Mol Med. 1999 May;3(5):473-6. doi: 10.3892/ijmm.3.5.473. Int J Mol Med. 1999. PMID: 10202177 Review.
Cited by
-
An analog of camptothecin inactive against Topoisomerase I is broadly neutralizing of HIV-1 through inhibition of Vif-dependent APOBEC3G degradation.Antiviral Res. 2016 Dec;136:51-59. doi: 10.1016/j.antiviral.2016.11.001. Epub 2016 Nov 5. Antiviral Res. 2016. PMID: 27825797 Free PMC article.
-
HIV-1 Vif interaction with APOBEC3 deaminases and its characterization by a new sensitive assay.J Neuroimmune Pharmacol. 2011 Jun;6(2):296-307. doi: 10.1007/s11481-011-9258-7. Epub 2011 Jan 29. J Neuroimmune Pharmacol. 2011. PMID: 21279453
-
The unique structure of the highly conserved PPLP region in HIV-1 Vif is critical for the formation of APOBEC3 recognition interfaces.mBio. 2025 Mar 12;16(3):e0333224. doi: 10.1128/mbio.03332-24. Epub 2025 Jan 21. mBio. 2025. PMID: 39835817 Free PMC article.
-
On the solution conformation and dynamics of the HIV-1 viral infectivity factor.J Mol Biol. 2011 Jul 29;410(5):1008-22. doi: 10.1016/j.jmb.2011.04.053. J Mol Biol. 2011. PMID: 21763503 Free PMC article.
-
Hydrodynamic and functional analysis of HIV-1 Vif oligomerization.Biochemistry. 2012 Mar 13;51(10):2078-86. doi: 10.1021/bi201738a. Epub 2012 Mar 5. Biochemistry. 2012. PMID: 22369580 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

