Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner
- PMID: 11071920
- PMCID: PMC15050
- DOI: 10.1091/mbc.11.11.3963
Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner
Abstract
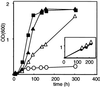
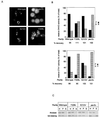
A number of peroxisome-associated proteins have been described that are involved in the import of proteins into peroxisomes, among which is the receptor for peroxisomal targeting signal 1 (PTS1) proteins Pex5p, the integral membrane protein Pex13p, which contains an Src homology 3 (SH3) domain, and the peripheral membrane protein Pex14p. In the yeast Saccharomyces cerevisiae, both Pex5p and Pex14p are able to bind Pex13p via its SH3 domain. Pex14p contains the classical SH3 binding motif PXXP, whereas this sequence is absent in Pex5p. Mutation of the conserved tryptophan in the PXXP binding pocket of Pex13-SH3 abolished interaction with Pex14p, but did not affect interaction with Pex5p, suggesting that Pex14p is the classical SH3 domain ligand and that Pex5p binds the SH3 domain in an alternative way. To identify the SH3 binding site in Pex5p, we screened a randomly mutagenized PEX5 library for loss of interaction with Pex13-SH3. Such mutations were all located in a small region in the N-terminal half of Pex5p. One of the altered residues (F208) was part of the sequence W(204)XXQF(208), that is conserved between Pex5 proteins of different species. Site-directed mutagenesis of Trp204 confirmed the essential role of this motif in recognition of the SH3 domain. The Pex5p mutants could only partially restore PTS1-protein import in pex5Delta cells in vivo. In vitro binding studies showed that these Pex5p mutants failed to interact with Pex13-SH3 in the absence of Pex14p, but regained their ability to bind in the presence of Pex14p, suggesting the formation of a heterotrimeric complex consisting of Pex5p, Pex14p, and Pex13-SH3. In vivo, these Pex5p mutants, like wild-type Pex5p, were still found to be associated with peroxisomes. Taken together, this indicates that in the absence of Pex13-SH3 interaction, other protein(s) is able to bind Pex5p at the peroxisome; Pex14p is a likely candidate for this function.
Figures







References
-
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JAKW, Veenhuis M, Kunau W-H. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. - PubMed
-
- Arold S, O'Brien R, Franken P, Strub M-P, Hoh F, Dumas C, Ladbury JE. RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry. 1998;37:14683–14691. - PubMed
-
- Bradford MM. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Brocard C, Kragler F, Simon MM, Schuster T, Hartig A. The tetratricopeptide repeat-domain of the PAS10 protein of Saccharomyces cerevisiae is essential for binding the peroxisomal targeting signal-SKL. Biochem Biophys Res Commun. 1994;204:1016–1022. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

