The microarray explorer tool for data mining of cDNA microarrays: application for the mammary gland
- PMID: 11071932
- PMCID: PMC113879
- DOI: 10.1093/nar/28.22.4452
The microarray explorer tool for data mining of cDNA microarrays: application for the mammary gland
Abstract
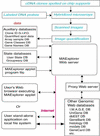
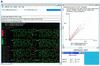
The Microarray Explorer (MAExplorer) is a versatile Java-based data mining bioinformatic tool for analyzing quantitative cDNA expression profiles across multiple microarray platforms and DNA labeling systems. It may be run as either a stand-alone application or as a Web browser applet over the Internet. With this program it is possible to (i) analyze the expression of individual genes, (ii) analyze the expression of gene families and clusters, (iii) compare expression patterns and (iv) directly access other genomic databases for clones of interest. Data may be downloaded as required from a Web server or in the case of the stand-alone version, reside on the user's computer. Analyses are performed in real-time and may be viewed and directly manipulated in images, reports, scatter plots, histograms, expression profile plots and cluster analyses plots. A key feature is the clone data filter for constraining a working set of clones to those passing a variety of user-specified logical and statistical tests. Reports may be generated with hypertext Web access to UniGene, GenBank and other Internet databases for sets of clones found to be of interest. Users may save their explorations on the Web server or local computer and later recall or share them with other scientists in this groupware Web environment. The emphasis on direct manipulation of clones and sets of clones in graphics and tables provides a high level of interaction with the data, making it easier for investigators to test ideas when looking for patterns. We have used the MAExplorer to profile gene expression patterns of 1500 duplicated genes isolated from mouse mammary tissue. We have identified genes that are preferentially expressed during pregnancy and during lactation. One gene we identified, carbonic anhydrase III, is highly expressed in mammary tissue from virgin and pregnant mice and in gene knock-out mice with underdeveloped mammary epithelium. Other genes, which include those encoding milk proteins, are preferentially expressed during lactation.
Figures


References
-
- Tukey J. (1977) Exploratory Data Analysis. Addison-Wesley Pub. Co., Reading, MA, pp. 1–688.
-
- Tufte E. (1997) Visual Explanations. Images and Quantities, Evidence and Narrative. Graphics Press, Cheshire, CT, pp. 1–156.
-
- Cleveland W.S. (1985) The Elements of Graphing Data. Wadsworth Press, Monterey, CA, pp. 1–323.
-
- Ermolaeva O., Rastogi,M., Pruitt,K.D., Schuler,G.D., Bittner,M.L., Chen,Y., Simon,R., Meltzer,P., Trent,J.M. and Boguski,M.S. (1998) Nature Genet., 20, 19–23. - PubMed
-
- Lipkin L.E. and Lemkin,P.F. (1980) Clin. Chem., 26, 1403–1412. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

