A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans
- PMID: 11071951
- PMCID: PMC113889
- DOI: 10.1093/nar/28.22.e97
A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans
Abstract
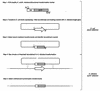
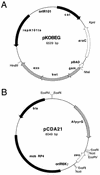
The construction of mutant fungal strains is often limited by the poor efficiency of homologous recombination in these organisms. Higher recombination efficiencies can be obtained by increasing the length of homologous DNA flanking the transformation marker, although this is a tedious process when standard molecular biology techniques are used for the construction of gene replacement cassettes. Here, we present a two-step technology which takes advantage of an Escherichia coli strain expressing the phage lambda Red(gam, bet, exo) functions and involves (i) the construction in this strain of a recombinant cosmid by in vivo recombination between a cosmid carrying a genomic region of interest and a PCR-generated transformation marker flanked by 50 bp regions of homology with the target DNA and (ii) genetic exchange in the fungus itself between the chromosomal locus and the circular or linearized recombinant cosmid. This strategy enables the rapid establishment of mutant strains carrying gene knock-outs with efficiencies >50%. It should also be appropriate for the construction of fungal strains with gene fusions or promoter replacements.
Figures

References
-
- van den Hondel C.A.M.J.J. and Punt,P.J. (1991) In Peberdy,J.F., Caten,C.E., Ogden,J.E. and Bennett,J.W. (eds), Applied Molecular Genetics of Fungi. Cambridge University Press, Cambridge, UK, pp. 1–28.
-
- d’Enfert C., Weidner,G. and Brakhage,A.A. (1999) In Brakhage,A.A., Jahn,B. and Schmidt,A. (eds), Aspergillus fumigatus: Biology, Clinics and Molecular Approaches to Pathogenicity. S. Karger AG, Basel, Switzerland, Vol. 2, pp. 149–166.
-
- Zhang Y., Buchholz,F., Muyrers,J.P.P. and Stewart,A.F. (1998) Nature Genet., 20, 123–128. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

