Peptide nucleic acids: versatile tools for gene therapy strategies
- PMID: 11072107
- PMCID: PMC4403640
- DOI: 10.1016/s0169-409x(00)00087-9
Peptide nucleic acids: versatile tools for gene therapy strategies
Abstract
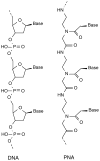
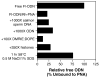
Peptide nucleic acids, or PNAs, are oligonucleotide analogs in which the phosphodiester backbone is replaced with a polyamide structure. First synthesized less than 10 years ago, they have received great attention due to their several favorable properties, including resistance to nuclease and protease digestion, stability in serum and cell extracts, and their high affinity for RNA and single and double-stranded DNA targets. Although initially designed and demonstrated to function as antisense and antigene reagents that inhibit both transcription and translation by steric hindrance, more recent applications have included gene activation by synthetic promoter formation and mutagenesis of chromosomal targets. Most notably for gene delivery, they have been used to specifically label plasmids and act as adapters to link synthetic peptides or ligands to the DNA. Thus, their great potential lies in the ability to attach specific targeting peptides to plasmids to circumvent such barriers to gene transfer as cell-targeting or nuclear localization, thereby increasing the efficacy of gene therapy.
Figures

References
-
- Nielsen PE, Eghoim M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. - PubMed
-
- Brown SC, Thomson SA, Veal JM, Davis DG. NMIR solution structure of a peptide nucleic acid complexed with RNA. Science. 1994;265:777–780. - PubMed
-
- Wittung P, Nielsen PE, Buchardt O, Eghoim M, Norden B. DNA-like double helix formed by peptide nucleic acid. Nature. 1994;368:561–563. - PubMed
-
- Egholm M, Buchardt O, Christensen L, Bebrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature. 1993;365:566–568. - PubMed
-
- Nielsen PE, Egholm M, Buchardt O. Evidence for (PNA)2IDNA triplex structure upon binding of PNA to dsDNA by strand displacement. J Mol Recognit. 1994;7:165–170. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

