Conversion of 4-hydroxyacetophenone into 4-phenyl acetate by a flavin adenine dinucleotide-containing Baeyer-Villiger-type monooxygenase
- PMID: 11073896
- PMCID: PMC111394
- DOI: 10.1128/JB.182.23.6565-6569.2000
Conversion of 4-hydroxyacetophenone into 4-phenyl acetate by a flavin adenine dinucleotide-containing Baeyer-Villiger-type monooxygenase
Abstract
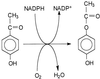
An arylketone monooxygenase was purified from Pseudomonas putida JD1 by ion exchange and affinity chromatography. It had the characteristics of a Baeyer-Villiger-type monooxygenase and converted its substrate, 4-hydroxyacetophenone, into 4-hydroxyphenyl acetate with the consumption of one molecule of oxygen and oxidation of one molecule of NADPH per molecule of substrate. The enzyme was a monomer with an M(r) of about 70,000 and contained one molecule of flavin adenine dinucleotide (FAD). The enzyme was specific for NADPH as the electron donor, and spectral studies showed rapid reduction of the FAD by NADPH but not by NADH. Other arylketones were substrates, including acetophenone and 4-hydroxypropiophenone, which were converted into phenyl acetate and 4-hydroxyphenyl propionate, respectively. The enzyme displayed Michaelis-Menten kinetics with apparent K(m) values of 47 microM for 4-hydroxyacetophenone, 384 microM for acetophenone, and 23 microM for 4-hydroxypropiophenone. The apparent K(m) value for NADPH with 4-hydroxyacetophenone as substrate was 17.5 microM. The N-terminal sequence did not show any similarity to other proteins, but an internal sequence was very similar to part of the proposed NADPH binding site in the Baeyer-Villiger monooxygenase cyclohexanone monooxygenase from an Acinetobacter sp.
Figures


Similar articles
-
4-Hydroxyacetophenone monooxygenase from Pseudomonas fluorescens ACB. A novel flavoprotein catalyzing Baeyer-Villiger oxidation of aromatic compounds.Eur J Biochem. 2001 May;268(9):2547-57. doi: 10.1046/j.1432-1327.2001.02137.x. Eur J Biochem. 2001. PMID: 11322873
-
Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-Δ(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.Appl Environ Microbiol. 2012 Apr;78(7):2200-12. doi: 10.1128/AEM.07694-11. Epub 2012 Jan 20. Appl Environ Microbiol. 2012. PMID: 22267661 Free PMC article.
-
Purification and characterization of cyclohexanone 1,2-monooxygenase from Exophiala jeanselmei strain KUFI-6N.Biosci Biotechnol Biochem. 2000 Dec;64(12):2696-8. doi: 10.1271/bbb.64.2696. Biosci Biotechnol Biochem. 2000. PMID: 11210139
-
Characterised Flavin-Dependent Two-Component Monooxygenases from the CAM Plasmid of Pseudomonas putida ATCC 17453 (NCIMB 10007): ketolactonases by Another Name.Microorganisms. 2018 Dec 20;7(1):1. doi: 10.3390/microorganisms7010001. Microorganisms. 2018. PMID: 30577535 Free PMC article. Review.
-
The Isoenzymic Diketocamphane Monooxygenases of Pseudomonas putida ATCC 17453-An Episodic History and Still Mysterious after 60 Years.Microorganisms. 2021 Dec 15;9(12):2593. doi: 10.3390/microorganisms9122593. Microorganisms. 2021. PMID: 34946195 Free PMC article. Review.
Cited by
-
Anaerobic degradation of p-ethylphenol by "Aromatoleum aromaticum" strain EbN1: pathway, regulation, and involved proteins.J Bacteriol. 2008 Aug;190(16):5699-709. doi: 10.1128/JB.00409-08. Epub 2008 Jun 6. J Bacteriol. 2008. PMID: 18539747 Free PMC article.
-
Proline dehydrogenase, a rate-limiting catabolic enzyme, affecting the growth and pathogenicity of Toxoplasma gondii tachyzoites by regulating the proline metabolism and mitochondrial function of the parasite.Parasit Vectors. 2025 Jul 29;18(1):309. doi: 10.1186/s13071-025-06966-x. Parasit Vectors. 2025. PMID: 40731026 Free PMC article.
-
The missing link in linear alkylbenzenesulfonate surfactant degradation: 4-sulfoacetophenone as a transient intermediate in the degradation of 3-(4-sulfophenyl)butyrate by Comamonas testosteroni KF-1.Appl Environ Microbiol. 2010 Jan;76(1):196-202. doi: 10.1128/AEM.02181-09. Epub 2009 Nov 13. Appl Environ Microbiol. 2010. PMID: 19915037 Free PMC article.
-
Cloning and characterization of a gene cluster involved in cyclopentanol metabolism in Comamonas sp. strain NCIMB 9872 and biotransformations effected by Escherichia coli-expressed cyclopentanone 1,2-monooxygenase.Appl Environ Microbiol. 2002 Nov;68(11):5671-84. doi: 10.1128/AEM.68.11.5671-5684.2002. Appl Environ Microbiol. 2002. PMID: 12406764 Free PMC article.
-
Substrate specificity and enantioselectivity of 4-hydroxyacetophenone monooxygenase.Appl Environ Microbiol. 2003 Jan;69(1):419-26. doi: 10.1128/AEM.69.1.419-426.2003. Appl Environ Microbiol. 2003. PMID: 12514023 Free PMC article.
References
-
- Britton L N, Markovetz A J. A novel ketone monooxygenase from Pseudomonas cepacia. J Biol Chem. 1977;252:8561–8566. - PubMed
-
- Cripps R E, Trudgill P W, Whateley J G. The metabolism of 1-phenylethanol and acetophenone by Nocardia T5 and an Arthrobacter species. Eur J Biochem. 1978;86:175–186. - PubMed
-
- Darby J M, Taylor D G, Hopper D J. Hydroquinone as the ring-fission substrate in the catabolism of 4-ethylphenol and 4-hydroxyacetophenone by Pseudomonas putida JD1. J Gen Microbiol. 1987;133:2137–2146.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

