InvB is a type III secretion chaperone specific for SspA
- PMID: 11073906
- PMCID: PMC111404
- DOI: 10.1128/JB.182.23.6638-6644.2000
InvB is a type III secretion chaperone specific for SspA
Abstract
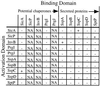
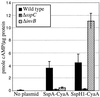
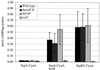
A wide variety of gram-negative bacteria utilize a specialized apparatus called the type III secretion system (TTSS) to translocate virulence factors directly into the cytoplasm of eukaryotic cells. These translocated effectors contribute to the pathogen's ability to infect and replicate within plant and animal hosts. The amino terminus of effector proteins contains sequences that are necessary and sufficient for both secretion and translocation by TTSS. Portions of these sequences contain binding sites for type III chaperones, which facilitate efficient secretion and translocation of specific effectors through TTSS. In this study, we have utilized the yeast two-hybrid assay to identify protein-protein interactions between effector and chaperone proteins encoded within Salmonella pathogenicity island 1 (SPI-1). Several interactions were identified including a novel interaction between the effector protein, SspA (SipA), and a putative chaperone, InvB. InvB was demonstrated to bind to the amino terminus of SspA in the bacterial cytoplasm. Furthermore, InvB acts as a type III chaperone for the efficient secretion and translocation of SspA by SPI-1. InvB also permitted translocation of SspA through the SPI-2 TTSS, indicating that it is an important regulator in the recognition of SspA as a target of TTSS. Finally, it was determined that InvB does not alter the transcription of sspA but that its absence results in reduced SspA protein levels in Salmonella enterica serovar Typhimurium.
Figures

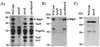

References
-
- Anderson D M, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. - PubMed
-
- Bartel P L, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. - PubMed
-
- Bennett J C, Hughes C. From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 2000;8:202–204. - PubMed
-
- Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

