Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6
- PMID: 11073908
- PMCID: PMC111406
- DOI: 10.1128/JB.182.23.6651-6658.2000
Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6
Abstract
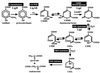
Protocatechuate (PCA) is the key intermediate metabolite in the lignin degradation pathway of Sphingomonas paucimobilis SYK-6 and is metabolized to pyruvate and oxaloacetate via the PCA 4,5-cleavage pathway. We characterized the 4-carboxy-2-hydroxymuconate-6-semialdehyde (CHMS) dehydrogenase gene (ligC). CHMS is the 4,5-cleavage product of PCA and is converted into 2-pyrone-4,6-dicarboxylate (PDC) by LigC. We found that ligC was located 295 bp downstream of ligB, which encodes the large subunit of the PCA 4,5-dioxygenase. The ligC gene consists of a 945-bp open reading frame encoding a polypeptide with a molecular mass of 34,590 Da. The deduced amino acid sequence of ligC showed 19 to 20% identity with 3-chlorobenzoate cis-dihydrodiol dehydrogenase of Alcaligenes sp. strain BR60 and phthalate cis-dihydrodiol dehydrogenases of Pseudomonas putida NMH102-2 and Burkholderia cepacia DBO1, which are unrelated to group I, II, and III microbial alcohol dehydrogenases (M. F. Reid and C. A. Fewson, Crit. Rev. Microbiol. 20:13-56, 1994). The ligC gene was expressed in Escherichia coli and LigC was purified to near homogeneity. Production of PDC from CHMS catalyzed by LigC was confirmed in the presence of NADP(+) by electrospray ionization-mass spectrometry and gas chromatography-mass spectrometry. LigC is a homodimer. The isoelectric point, optimum pH, and optimum temperature were estimated to be 5.3, 8.0, and 25 degrees C, respectively. The K(m) for NADP(+) was estimated to be 24.6 +/- 1.5 microM, which was approximately 10 times lower than that for NAD(+) (252 +/- 3.9 microM). The K(m)s for CHMS in the presence of NADP(+) and NAD(+) are 26.0 +/- 0.5 and 20.6 +/- 1.0 microM, respectively. Disruption of ligC in S. paucimobilis SYK-6 prevented growth with vanillate. Only PCA was accumulated during the incubation of vanillate with the whole cells of the ligC insertion mutant (DLC), indicating a lack of PCA 4,5-dioxygenase activity in DLC. However, the introduction of ligC into DLC restored its ability to grow on vanillate. PDC was suggested to be an inducer for ligAB gene expression.
Figures



References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990.
-
- Bagdasarian M, Lurz R, Rückert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010 derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. - PubMed
-
- Bolivar F, Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

