Crk1, a novel Cdc2-related protein kinase, is required for hyphal development and virulence in Candida albicans
- PMID: 11073971
- PMCID: PMC86484
- DOI: 10.1128/MCB.20.23.8696-8708.2000
Crk1, a novel Cdc2-related protein kinase, is required for hyphal development and virulence in Candida albicans
Abstract
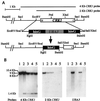
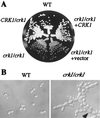
Both mitogen-activated protein kinases and cyclin-dependent kinases play a role in hyphal development in Candida albicans. Using an oligonucleotide probe-based screen, we have isolated a new member of the Cdc2 kinase subfamily, designated Crk1 (Cdc2-related kinase). The protein sequence of Crk1 is most similar to those of Saccharomyces cerevisiae Sgv1 and human Pkl1/Cdk9. In S. cerevisiae, CRK1 suppresses some, but not all, of the defects associated with an sgv1 mutant. Deleting both copies of CRK1 in C. albicans slows growth slightly but leads to a profound defect in hyphal development under all conditions examined. crk1/crk1 mutants are impaired in the induction of hypha-specific genes and are avirulent in mice. Consistent with this, ectopic expression of the Crk1 kinase domain (CRK1N) promotes filamentous or invasive growth in S. cerevisiae and hyphal development in C. albicans. The activity of Crk1 in S. cerevisiae requires Flo8 but is independent of Ste12 and Phd1. Similarly, Crk1 promotes filamentation through a route independent of Cph1 and Efg1 in C. albicans. RAS1(V13) can also activate filamentation in a cph1/cph1 efg1/efg1 double mutant. Interestingly, CRK1N produces florid hyphae in ras1/ras1 strains, while RAS1(V13) generates feeble hyphae in crk1/crk1 strains.
Figures









References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
