In vivo requirement of activator-specific binding targets of mediator
- PMID: 11073972
- PMCID: PMC86488
- DOI: 10.1128/MCB.20.23.8709-8719.2000
In vivo requirement of activator-specific binding targets of mediator
Abstract
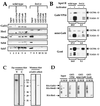
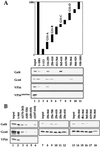
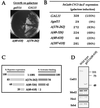
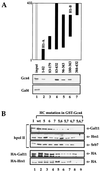
There has been no unequivocal demonstration that the activator binding targets identified in vitro play a key role in transcriptional activation in vivo. To examine whether activator-Mediator interactions are required for gene transcription under physiological conditions, we performed functional analyses with Mediator components that interact specifically with natural yeast activators. Different activators interact with Mediator via distinct binding targets. Deletion of a distinct activator binding region of Mediator completely compromised gene activation in vivo by some, but not all, transcriptional activators. These demonstrate that the activator-specific targets in Mediator are essential for transcriptional activation in living cells, but their requirement was affected by the nature of the activator-DNA interaction and the existence of a postrecruitment activation process.
Figures



References
-
- Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. - PubMed
-
- Björklund S, Kim Y-J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. - PubMed
-
- Brown C E, Lechner T, Howe L, Workman J L. The many HATs of transcription coactivators. Trends Biochem Sci. 2000;25:15–19. - PubMed
-
- Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
