Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1
- PMID: 11073977
- PMCID: PMC86503
- DOI: 10.1128/MCB.20.23.8758-8766.2000
Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1
Abstract
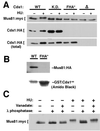
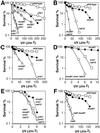
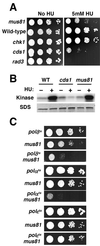
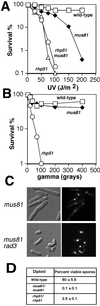
Cds1, a serine/threonine kinase, enforces the S-M checkpoint in the fission yeast Schizosaccharomyces pombe. Cds1 is required for survival of replicational stress caused by agents that stall replication forks, but how Cds1 performs these functions is largely unknown. Here we report that the forkhead-associated-1 (FHA1) protein-docking domain of Cds1 interacts with Mus81, an evolutionarily conserved damage tolerance protein. Mus81 has an endonuclease homology domain found in the XPF nucleotide excision repair protein. Inactivation of mus81 reveals a unique spectrum of phenotypes. Mus81 enables survival of deoxynucleotide triphosphate starvation, UV radiation, and DNA polymerase impairment. Mus81 is essential in the absence of Bloom's syndrome Rqh1 helicase and is required for productive meiosis. Genetic epistasis studies suggest that Mus81 works with recombination enzymes to properly replicate damaged DNA. Inactivation of Mus81 triggers a checkpoint-dependent delay of mitosis. We propose that Mus81 is involved in the recruitment of Cds1 to aberrant DNA structures where Cds1 modulates the activity of damage tolerance enzymes.
Figures

References
-
- Bahler J, Wu J, Longtine M S, Shah N G, McKenzie A, Steever A B, Wach A, Phileppsen P, Pringle J R. Heterologous modules for efficient and versatile PCR-based gene targetting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Bell D W, Varley J M, Szydlo T E, Kang D H, Wahrer D C, Shannon K E, Lubratovich M, Verselis S J, Isselbacher K J, Fraumeni J F, Birch J M, Li F P, Garber J E, Haber D A. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
