Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy
- PMID: 11076515
- PMCID: PMC2903970
- DOI: 10.1021/bi001472t
Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy
Abstract
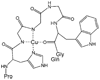
Recent evidence indicates that the prion protein (PrP) plays a role in copper metabolism in the central nervous system. The N-terminal region of human PrP contains four sequential copies of the highly conserved octarepeat sequence PHGGGWGQ spanning residues 60-91. This region selectively binds divalent copper ions (Cu(2+)) in vivo. To elucidate the specific mode and site of binding, we have studied a series of Cu(2+)-peptide complexes composed of 1-, 2-, and 4-octarepeats and several sub-octarepeat peptides, by electron paramagnetic resonance (EPR, conventional X-band and low-frequency S-band) and circular dichroism (CD) spectroscopy. At pH 7.45, two EPR active binding modes are observed where the dominant mode appears to involve coordination of three nitrogens and one oxygen to the copper ion, while in the minor mode two nitrogens and two oxygens coordinate. ESEEM spectra demonstrate that the histidine imidazole contributes one of these nitrogens. The truncated sequence HGGGW gives EPR and CD that are indistinguishable from the dominant binding mode observed for the multi-octarepeat sequences and may therefore comprise the fundamental Cu(2+) binding unit. Both EPR and CD titration experiments demonstrate rigorously a 1:1 Cu(2+)/octarepeat binding stoichiometry regardless of the number of octarepeats in a given peptide sequence. Detailed spin integration of the EPR signals demonstrates that all of the bound Cu(2+) is detected thereby ruling out strong exchange coupling that is often found when there is imidazolate bridging between paramagnetic metal centers. A model consistent with these data is proposed in which Cu(2+) is bound to the nitrogen of the histidine imidazole side chain and to two nitrogens from sequential glycine backbone amides.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

