Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation
- PMID: 11076856
- PMCID: PMC310985
- DOI: 10.1101/gr.161600
Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation
Abstract
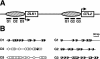
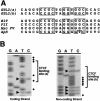
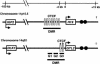
The evolution of genomic imprinting in mammals occurred more than 100 million years ago, and resulted in the formation of genes that are functionally haploid because of parent-of-origin-dependent expression. Despite ample evidence from studies in a number of species suggesting the presence of imprinted genes on human chromosome 14, their identity has remained elusive. Here we report the identification of two reciprocally imprinted genes, GTL2 and DLK1, which together define a novel imprinting cluster on human chromosome 14q32. The maternally expressed GTL2 (gene trap locus 2) gene encodes for a nontranslated RNA. DLK1 (delta, Drosophila, homolog-like 1) is a paternally expressed gene that encodes for a transmembrane protein containing six epidermal growth factor (EGF) repeat motifs closely related to those present in the delta/notch/serrate family of signaling molecules. The paternal expression, chromosomal localization, and biological function of DLK1 also make it a likely candidate gene for the callipyge phenotype in sheep. Many of the predicted structural and regulatory features of the DLK1/GTL2 domain are highly analogous to those implicated in IGF2/H19 imprint regulation, including two hemimethylated consensus binding sites for the vertebrate enhancer blocking protein, CTCF. These results provide evidence that a common mechanism and domain organization may be used for juxtapositioned, reciprocally imprinted genes.
Figures


Comment in
-
Comparative genomics sheds light on mechanisms of genomic imprinting.Genome Res. 2000 Nov;10(11):1660-3. doi: 10.1101/gr.166200. Genome Res. 2000. PMID: 11076850 Review. No abstract available.
References
-
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. - PubMed
-
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. - PubMed
-
- Carlsson C, Tornehave D, Lindberg K, Galante P, Billestrup N, Michelsen B, Larsson LI, Nielsen JH. Growth hormone and prolactin stimulate the expression of rat preadipocyte factor-1/delta-like protein in pancreatic islets: Molecular cloning and expression pattern during development and growth of the endocrine pancreas. Endocrinology. 1997;138:3940–3948. - PubMed
-
- Cattanach BM, Beechey CV. Genomic imprinting in the mouse: Possible final analysis. In: Reik W, Surani A, editors. Genomic Imprinting. Oxford, UK: IRL Press; 1997. pp. 118–145.
-
- Cockett NE, Jackson SP, Shay TL, Farnir F, Berghmans S, Snowder GD, Nielsen DM, Georges M. Polar overdominance at the ovine callipyge locus. Science. 1996;273:236–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
