Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors
- PMID: 11078516
- PMCID: PMC27210
- DOI: 10.1073/pnas.230442897
Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors
Abstract
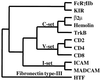
Paired Ig-like receptors (PIR) that can reciprocally modulate cellular activation have been described in mammals. In the present study, we searched expressed sequence tag databases for PIR relatives to identify chicken expressed sequence tags predictive of approximately 25% amino acid identity to mouse PIR. Rapid amplification of cDNA ends (RACE)-PCR extension of expressed sequence-tag sequences using chicken splenic cDNA as a template yielded two distinct cDNAs, the sequence analysis of which predicted protein products with related extracellular Ig-like domains. Chicken Ig-like receptor (CHIR)-A was characterized by its transmembrane segment with a positively charged histidine residue and short cytoplasmic tail, thereby identifying CHIR-A as a candidate-activating receptor. Conversely, CHIR-B was characterized by its nonpolar transmembrane segment and cytoplasmic tail with two immunoreceptor tyrosine-based inhibitory motifs, indicating that it may serve as an inhibitory receptor. The use of CHIR amino acid sequences in a search for other PIR relatives led to the recognition of mammalian Fc receptors as distantly related genes. Comparative analyses based on amino acid sequences and three-dimensional protein structures provided molecular evidence for common ancestry of the PIR and Fc receptor gene families.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

