A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR)
- PMID: 11078525
- PMCID: PMC27207
- DOI: 10.1073/pnas.240444197
A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR)
Abstract
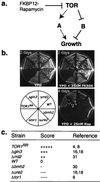
The target of rapamycin protein (TOR) is a highly conserved ataxia telangiectasia-related protein kinase essential for cell growth. Emerging evidence indicates that TOR signaling is highly complex and is involved in a variety of cellular processes. To understand its general functions, we took a chemical genomics approach to explore the genetic interaction between TOR and other yeast genes on a genomic scale. In this study, the rapamycin sensitivity of individual deletion mutants generated by the Saccharomyces Genome Deletion Project was systematically measured. Our results provide a global view of the rapamycin-sensitive functions of TOR. In contrast to conventional genetic analysis, this approach offers a simple and thorough analysis of genetic interaction on a genomic scale and measures genetic interaction at different possible levels. It can be used to study the functions of other drug targets and to identify novel protein components of a conserved core biological process such as DNA damage checkpoint/repair that is interfered with by a cell-permeable chemical compound.
Figures
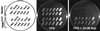
References
-
- Dennis P B, Fumagalli S, Thomas G. Curr Opin Genet Dev. 1999;9:49–54. - PubMed
-
- Kurnuvilla F, Schreiber S L. Chem Biol. 1999;6:R129–R136. - PubMed
-
- Heitman J, Movva N R, Hall M N. Science. 1991;253:905–909. - PubMed
-
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Cell. 1993;73:585–596. - PubMed
-
- Cafferkey R, McLaughlin M M, Young P R, Johnson R K, Livi G P. Gene. 1994;141:133–136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

