Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4
- PMID: 11080155
- PMCID: PMC305831
- DOI: 10.1093/emboj/19.22.6085
Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4
Abstract
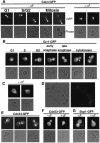
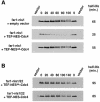
Far1 is a bifunctional protein that is required to arrest the cell cycle and establish cell polarity during yeast mating. Here we show that SCF(Cdc4) ubiquitylates Far1 in the nucleus, which in turn targets the multi-ubiquitylated protein to 26S proteasomes most likely located at the nuclear envelope. In response to mating pheromones, a fraction of Far1 was stabilized after its export into the cytoplasm by Ste21/Msn5. Preventing nuclear export destabilized Far1, while conversely cytoplasmic Far1 was stabilized, although the protein was efficiently phosphorylated in a Cdc28-Cln-dependent manner. The core SCF subunits Cdc53, Hrt1 and Skp1 were distributed in the nucleus and the cytoplasm, whereas the F-box protein Cdc4 was exclusively nuclear. A cytoplasmic form of Cdc4 was unable to complement the growth defect of cdc4-1 cells, but it was sufficient to degrade Far1 in the cytoplasm. Our results illustrate the importance of subcellular localization of F-box proteins, and provide an example of how an extracellular signal regulates protein stability at the level of substrate localization.
Figures








References
-
- Bai C., Sen,P., Hofmann,K., Ma,L., Goebl,M., Harper,J.W. and Elledge,S.J. (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell, 86, 263–274. - PubMed
-
- Barral Y., Jentsch,S. and Mann,C. (1995) G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev., 9, 399–409. - PubMed
-
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

