Modular domain structure of Arabidopsis COP1. Reconstitution of activity by fragment complementation and mutational analysis of a nuclear localization signal in planta
- PMID: 11080276
- PMCID: PMC59198
- DOI: 10.1104/pp.124.3.979
Modular domain structure of Arabidopsis COP1. Reconstitution of activity by fragment complementation and mutational analysis of a nuclear localization signal in planta
Abstract
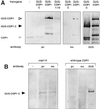
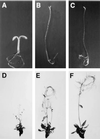
The Arabidopsis COP1 protein functions as a developmental regulator, in part by repressing photomorphogenesis in darkness. Using complementation of a cop1 loss-of-function allele with transgenes expressing fusions of cop1 mutant proteins and beta-glucuronidase, it was confirmed that COP1 consists of two modules, an amino terminal module conferring a basal function during development and a carboxyl terminal module conferring repression of photomorphogenesis. The amino-terminal zinc-binding domain of COP1 was indispensable for COP1 function. In contrast, the debilitating effects of site-directed mutations in the single nuclear localization signal of COP1 were partially compensated by high-level transgene expression. The carboxyl-terminal module of COP1, though unable to substantially ameliorate a cop1 loss-of-function allele on its own, was sufficient for conferring a light-quality-dependent hyperetiolation phenotype in the presence of wild-type COP1. Moreover, partial COP1 activity could be reconstituted in vivo from two non-covalently linked, complementary polypeptides that represent the two functional modules of COP1. Evidence is presented for efficient association of the two sub-fragments of the split COP1 protein in Arabidopsis and in a yeast two-hybrid assay.
Figures

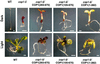


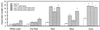

Similar articles
-
Short communication: the N-terminal fragment of Arabidopsis photomorphogenic repressor COP1 maintains partial function and acts in a concentration-dependent manner.Plant J. 1999 Dec;20(6):713-7. doi: 10.1046/j.1365-313x.1999.00639.x. Plant J. 1999. PMID: 10652143
-
Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis.Plant Physiol. 1997 Jul;114(3):779-88. doi: 10.1104/pp.114.3.779. Plant Physiol. 1997. PMID: 9232869 Free PMC article.
-
Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1.Plant Cell. 1999 Mar;11(3):349-64. doi: 10.1105/tpc.11.3.349. Plant Cell. 1999. PMID: 10072396 Free PMC article.
-
Structural organization and interactions of COP1, a light-regulated developmental switch.Plant Mol Biol. 1999 Sep;41(2):151-8. doi: 10.1023/a:1006324115086. Plant Mol Biol. 1999. PMID: 10579483 Review.
-
The role of COP1 in repression of Arabidopsis photomorphogenic development.Trends Cell Biol. 1999 Mar;9(3):113-8. doi: 10.1016/s0962-8924(99)01499-3. Trends Cell Biol. 1999. PMID: 10201077 Review.
Cited by
-
SPA Proteins Affect the Subcellular Localization of COP1 in the COP1/SPA Ubiquitin Ligase Complex during Photomorphogenesis.Plant Physiol. 2017 Jul;174(3):1314-1321. doi: 10.1104/pp.17.00488. Epub 2017 May 23. Plant Physiol. 2017. PMID: 28536102 Free PMC article.
-
Genetic interactions of Arabidopsis thaliana damaged DNA binding protein 1B (DDB1B) with DDB1A, DET1, and COP1.G3 (Bethesda). 2013 Mar;3(3):493-503. doi: 10.1534/g3.112.005249. Epub 2013 Mar 1. G3 (Bethesda). 2013. PMID: 23450167 Free PMC article.
-
OsCOP1 regulates embryo development and flavonoid biosynthesis in rice (Oryza sativa L.).Theor Appl Genet. 2021 Aug;134(8):2587-2601. doi: 10.1007/s00122-021-03844-9. Epub 2021 May 5. Theor Appl Genet. 2021. PMID: 33950284 Free PMC article.
-
Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time.Plant Cell. 2010 Jan;22(1):108-23. doi: 10.1105/tpc.109.065490. Epub 2010 Jan 8. Plant Cell. 2010. PMID: 20061554 Free PMC article.
-
The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer.Proc Natl Acad Sci U S A. 2004 Apr 27;101(17):6798-802. doi: 10.1073/pnas.0307964101. Epub 2004 Apr 14. Proc Natl Acad Sci U S A. 2004. PMID: 15084749 Free PMC article.
References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: New York Greene Publishing/Wiley Interscience; 1997.
-
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. - PubMed
-
- Deng X-W, Caspar T, Quail PH. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 1991;5:1172–1182. - PubMed
-
- Deng X-W, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a novel protein with both a Zn-binding motif and a Gβ homologous domain. Cell. 1992;71:791–801. - PubMed
-
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

