A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize
- PMID: 11080288
- PMCID: PMC59210
- DOI: 10.1104/pp.124.3.1105
A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize
Abstract
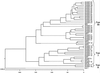
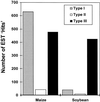
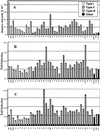
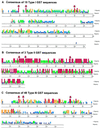
By BLAST searching a large expressed sequence tag database for glutathione S-transferase (GST) sequences we have identified 25 soybean (Glycine max) and 42 maize (Zea mays) clones and obtained accurate full-length GST sequences. These clones probably represent the majority of members of the GST multigene family in these species. Plant GSTs are divided according to sequence similarity into three categories: types I, II, and III. Among these GSTs only the active site serine, as well as another serine and arginine in or near the "G-site" are conserved throughout. Type III GSTs have four conserved sequence patches mapping to distinct structural features. Expression analysis reveals the distribution of GSTs in different tissues and treatments: Maize GSTI is overall the most highly expressed in maize, whereas the previously unknown GmGST 8 is most abundant in soybean. Using DNA microarray analysis we observed increased expression among the type III GSTs after inducer treatment of maize shoots, with different genes responding to different treatments. Protein activity for a subset of GSTs varied widely with seven substrates, and any GST exhibiting greater than marginal activity with chloro-2,4 dinitrobenzene activity also exhibited significant activity with all other substrates, suggesting broad individual enzyme substrate specificity.
Figures

References
-
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, McCombie WR, Venter JC. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Andrews CJ, Jepson I, Skipsey M, Townson JK, Edwards R. Nucleotide sequence of a glutathione transferase (accession no. Y10820) from soybean with activity towards herbicides. Plant Physiol. 1997;113:1005.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

