Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance
- PMID: 11083623
- PMCID: PMC90188
- DOI: 10.1128/AAC.44.12.3249-3256.2000
Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance
Figures
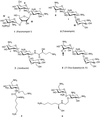
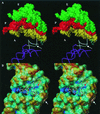
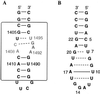
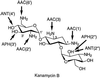
References
-
- Alper P B, Hendrix M, Sears P, Wong C-H. Probing the specificity of aminoglycoside-ribosomal RNA interactions with designed synthetic analogs. J Am Chem Soc. 1998;120:1965–1978.
-
- Azucena E, Grapsas I, Mobashery S. Properties of a bifunctional bacterial antibiotic resistance enzyme that catalyzes ATP-dependent 2"-phosphorylation and acetyl-CoA-dependent 6′-acetylation of aminoglycosides. J Am Chem Soc. 1997;119:2317–2318.
-
- Beauclerk A A, Cundliffe E. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J Mol Biol. 1987;193:661–671. - PubMed
-
- Cate J H, Yusupov M M, Yusupova G Z, Earnest T E, Noller H F. X-ray crystal structure of 70S ribosome functional complexes. Science. 1999;285:2095–2104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

