Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA
- PMID: 11083797
- PMCID: PMC97782
- DOI: 10.1128/IAI.68.12.6790-6797.2000
Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA
Abstract
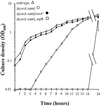
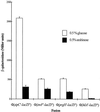
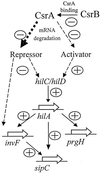
Penetration of intestinal epithelial cells by Salmonella enterica serovar Typhimurium requires the expression of invasion genes, found in Salmonella pathogenicity island 1 (SPI1), that encode components of a type III secretion apparatus. These genes are controlled in a complex manner by regulators within SPI1, including HilA and InvF, and those outside SPI1, such as the two-component regulators PhoP/PhoQ and BarA/SirA. We report here that epithelial cell invasion requires the serovar Typhimurium homologue of Escherichia coli csrA, which encodes a regulator that alters the stability of specific mRNA targets. A deletion mutant of csrA was unable to efficiently invade cultured epithelial cells and showed reduced expression of four tested SPI1 genes, hilA, invF, sipC, and prgH. Overexpression of csrA from an induced araBAD promoter also negatively affected the expression of these genes, indicating that CsrA can act as both a positive and a negative regulator of SPI1 genes and suggesting that the bacterium must tightly control the level or activity of CsrA to achieve maximal invasion. We found that CsrA affected hilA, a regulator of the other three genes we tested, probably by controlling one or more genetic elements that regulate hilA. We also found that both the loss and the overexpression of csrA reduced the expression of two regulators of hilA, hilC and hilD, suggesting that csrA exerts its control of hilA through one or both of these regulators. We further found, however, that CsrA could affect the expression of both invF and sipC independent of its effects on hilA. One additional striking phenotype of the csrA mutant, not observed in a comparable E. coli mutant, was its slow growth. Phenotypic revertants that had normal growth rates, while maintaining the csrA mutation, were common. These suppressed strains, however, did not recover the ability to invade cultured cells, indicating that the csrA-mediated loss of invasion cannot be attributed simply to poor growth and that the growth and invasion deficits of the csrA mutant arise from effects of CsrA on different targets.
Figures


References
-
- Ahmer B M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. - PubMed
-
- Altier C, Suyemoto M, Ruiz A I, Burnham K D, Maurer R. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol. 2000;35:635–646. - PubMed
-
- Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. - PubMed
-
- Bajaj V, Lucas R L, Hwang C, Lee C A. Coordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

