Chromosomal addresses of the cohesin component Mcd1p
- PMID: 11086006
- PMCID: PMC2174344
- DOI: 10.1083/jcb.151.5.1047
Chromosomal addresses of the cohesin component Mcd1p
Abstract
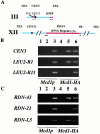
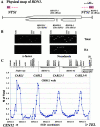
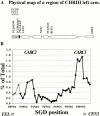
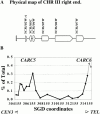
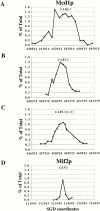
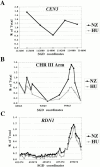
We identified the chromosomal addresses of a cohesin subunit, Mcd1p, in vivo by chromatin immunoprecipitation coupled with high resolution PCR-based chromosomal walking. The mapping of new Mcd1p-binding sites (cohesin-associated regions [CARs]) in single-copy sequences of several chromosomes establish their spacing ( approximately 9 kb), their sequestration to intergenic regions, and their association with AT-rich sequences as general genomic properties of CARs. We show that cohesins are not excluded from telomere proximal regions, and the enrichment of cohesins at the centromere at mitosis reflects de novo loading. The average size of a CAR is 0.8-1.0 kb. They lie at the boundaries of transcriptionally silenced regions, suggesting they play a direct role in defining the silent chromatin domain. Finally, we identify CARs in tandem (rDNA) and interspersed repetitive DNA (Ty2 and subtelomeric repeats). Each 9-kb rDNA repeat has a single CAR proximal to the 5S gene. Thus, the periodicity of CARs in single-copy regions and the rDNA repeats is conserved. The presence and spacing of CARs in repetitive DNA has important implications for genomic stability and chromosome packaging/condensation.
Figures


References
-
- Bajer A. Ciné-micrographic studies on chromosome movements in beta-irradiated cells. Chromosoma. 1958;9:319–331. - PubMed
-
- Bell S.P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. - PubMed
-
- Bickmore W.A., Oghene K. Visualizing the spatial relationships between defined DNA sequences and the axial region of extracted metaphase chromosomes. Cell. 1996;84:95–104. - PubMed
-
- Blat Y., Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. - PubMed
-
- Bryk M., Banerjee M., Murphy M., Knudsen K.E., Garfinkel D.J., Curcio M.J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

