Chromatin assembly at kinetochores is uncoupled from DNA replication
- PMID: 11086012
- PMCID: PMC2174364
- DOI: 10.1083/jcb.151.5.1113
Chromatin assembly at kinetochores is uncoupled from DNA replication
Abstract
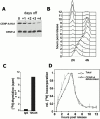
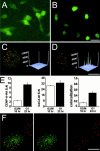
The specification of metazoan centromeres does not depend strictly on centromeric DNA sequences, but also requires epigenetic factors. The mechanistic basis for establishing a centromeric "state" on the DNA remains unclear. In this work, we have directly examined replication timing of the prekinetochore domain of human chromosomes. Kinetochores were labeled by expression of epitope-tagged CENP-A, which stably marks prekinetochore domains in human cells. By immunoprecipitating CENP-A mononucleosomes from synchronized cells pulsed with [(3)H]thymidine we demonstrate that CENP-A-associated DNA is replicated in mid-to-late S phase. Cytological analysis of DNA replication further demonstrated that centromeres replicate asynchronously in parallel with numerous other genomic regions. In contrast, quantitative Western blot analysis demonstrates that CENP-A protein synthesis occurs later, in G2. Quantitative fluorescence microscopy and transient transfection in the presence of aphidicolin, an inhibitor of DNA replication, show that CENP-A can assemble into centromeres in the absence of DNA replication. Thus, unlike most genomic chromatin, histone synthesis and assembly are uncoupled from DNA replication at the kinetochore. Uncoupling DNA replication from CENP-A synthesis suggests that regulated chromatin assembly or remodeling could play a role in epigenetic centromere propagation.
Figures


References
-
- Barry A.E., Howman E.V., Cancilla M.R., Saffery R., Choo K.H. Sequence analysis of an 80-kb human neocentromere. Hum. Mol. Genet. 1999;8:217–227. - PubMed
-
- Buchwitz B.J., Ahmad K., Moore L.L., Roth M.B., Henikoff S. A histone-H3-like protein in C. elegans . Nature. 1999;401:547–548. - PubMed
-
- Csink A.K., Henikoff S. Something from nothingthe evolution and utility of satellite repeats. Trends Genet. 1998;14:200–204. - PubMed
-
- Ekwall K., Olsson T., Turner B.M., Cranston G., Allshire R.C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. - PubMed

