Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension
- PMID: 11086013
- PMCID: PMC2174362
- DOI: 10.1083/jcb.151.5.1119
Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension
Abstract
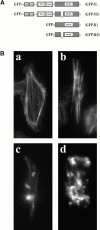
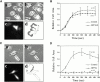
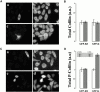
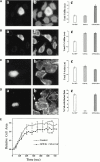
In metastatic rat mammary adenocarcinoma cells, cell motility can be induced by epidermal growth factor. One of the early events in this process is the massive generation of actin barbed ends, which elongate to form filaments immediately adjacent to the plasma membrane at the tip of the leading edge. As a result, the membrane moves outward and forms a protrusion. To test the involvement of ADF/cofilin in the stimulus-induced barbed end generation at the leading edge, we inhibited ADF/cofilin's activity in vivo by increasing its phosphorylation level using the kinase domain of LIM-kinase 1 (GFP-K). We report here that expression of GFP-K in rat cells results in the near total phosphorylation of ADF/cofilin, without changing either the G/F-actin ratio or signaling from the EGF receptor in vivo. Phosphorylation of ADF/cofilin is sufficient to completely inhibit the appearance of barbed ends and lamellipod protrusion, even in the continued presence of abundant G-actin. Coexpression of GFP-K, together with an active, nonphosphorylatable mutant of cofilin (S3A cofilin), rescues barbed end formation and lamellipod protrusion, indicating that the effects of kinase expression are caused by the phosphorylation of ADF/cofilin. These results indicate a direct role for ADF/cofilin in the generation of the barbed ends that are required for lamellipod extension in response to EGF stimulation.
Figures



References
-
- Arber S., Brbayannis F.A., Hanser H., Schneider C., Stanyon C.A., Bernard O., Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase 1. Nature. 1998;393:805–809. - PubMed
-
- Bailly M., Yan L., Whitesides G.M., Condeelis J.S., Segall J.E. Regulation of protrusion shape and adhesion to the substratum during chemotactic responses of mammalian carcinoma cells. Exp. Cell Res. 1998;241:285–299. - PubMed
-
- Bamburg J.R. Proteins of the ADF/Cofilin familyessential regulators of actin Dynamics. Annu. Rev. Cell. Dev. Biol. 1999;15:185–230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

