pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus
- PMID: 11087364
- PMCID: PMC3060174
- DOI: 10.1021/bi001799u
pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus
Abstract
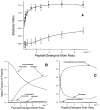
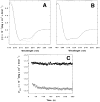
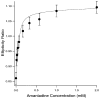
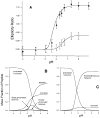
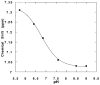
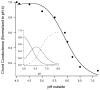
The M2 proton channel from the influenza A virus is a small protein with a single transmembrane helix that associates to form a tetramer in vivo. This protein forms proton-selective ion channels, which are the target of the drug amantadine. Here, we propose a mechanism for the pH-dependent association, and amantadine binding of M2, based on studies of a peptide representing the M2 transmembrane segment in dodecylphosphocholine micelles. Using analytical ultracentrifugation, we find that the sedimentation curves for the peptide depend on its concentration in the micellar phase. The data are well-described by a monomer-tetramer equilibrium, and the binding of amantadine shifts the monomer-tetramer equilibrium toward tetrameric species. Both tetramerization and the binding of amantadine lead to increases in the magnitude of the ellipticity at 223 nm in the circular dichroism spectrum of the peptide. The tetramerization and binding of amantadine are more favorable at elevated pH, with a pK(a) that is assigned to a His side chain, the only ionizable residue within the transmembrane helix. Our results, interpreted quantitatively in terms of a reversible monomer and tetramer protonation equilibrium model, suggest that amantadine competes with protons for binding to the deprotonated tetramer, thereby stabilizing the tetramer in a slightly altered conformation. This model accounts for the observed inhibition of proton flux by amantadine. Additionally, our measurements suggest that the M2 tetramer is substantially protonated at neutral pH and that both singly and doubly protonated states could be involved in M2's proton conduction at more acidic pHs.
Figures

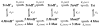
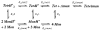
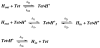
References
-
- Lamb RA, Holsinger LJ, Pinto LH. In: Receptor-Mediated Virus Entry into Cells. Wimmer E, editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. pp. 303–321.
-
- Duff KC, Kelly SM, Price NC, Bradshaw JP. FEBS Lett. 1992;311:256–258. - PubMed
-
- Duff KC, Gilchrist PJ, Saxena AM, Bradshaw JP. Virology. 1994;202:287–293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

