The zalpha domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA
- PMID: 11087828
- PMCID: PMC17610
- DOI: 10.1073/pnas.240464097
The zalpha domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA
Abstract
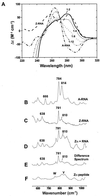
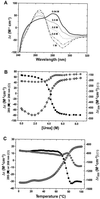
The Zalpha domain of human double-stranded RNA adenosine deaminase 1 binds specifically to left-handed Z-DNA and stabilizes the Z-conformation. Here we report spectroscopic and analytical results that demonstrate that Zalpha can also stabilize the left-handed Z-conformation in double-stranded RNA. Zalpha induces a slow transition from the right-handed A-conformation to the Z-form in duplex r(CG)(6), with an activation energy of 38 kcal mol(-1). We conclude that Z-RNA as well as Z-DNA can be accommodated in the tailored binding site of Zalpha. The specific binding of Z-RNA by Zalpha may be involved in targeting double-stranded RNA adenosine deaminase 1 for a role in hypermutation of RNA viruses.
Figures

References
-
- Higuchi M, Single F N, Kohler M, Sommer B, Sprengel R, Seeburg P H. Cell. 1993;75:1361–1370. - PubMed
-
- Schwartz T, Lowenhaupt K, Kim Y G, Li L, Brown B A, II, Herbert A, Rich A. J Biol Chem. 1999;274:2899–2906. - PubMed
-
- Schade M, Behlke J, Lowenhaupt K, Herbert A, Rich A, Oschkinat H. FEBS Lett. 1999;458:27–31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

