Protein folding and unfolding in microseconds to nanoseconds by experiment and simulation
- PMID: 11087839
- PMCID: PMC17607
- DOI: 10.1073/pnas.250473497
Protein folding and unfolding in microseconds to nanoseconds by experiment and simulation
Erratum in
- Proc Natl Acad Sci U S A 2001 Jan 16;98(2):777
Abstract
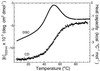
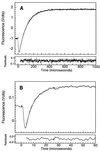
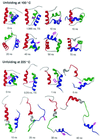
The Engrailed Homeodomain protein has the highest refolding and unfolding rate constants directly observed to date. Temperature jump relaxation measurements gave a refolding rate constant of 37,500 s(-1) in water at 25 degrees C, rising to 51,000 s(-1) around 42 degrees C. The unfolding rate constant was 1,100 s(-1) in water at 25 degrees C and 205,000 s(-1) at 63 degrees C. The unfolding half-life is extrapolated to be approximately 7.5 ns at 100 degrees C, which allows real-time molecular dynamics unfolding simulations to be tested on this system at a realistic temperature. Preliminary simulations did indeed conform to unfolding on this time scale. Further, similar transition states were observed in simulations at 100 degrees C and 225 degrees C, suggesting that high-temperature simulations provide results applicable to lower temperatures.
Figures

References
-
- Chamberlain A K, Marqusee S. Adv Protein Chem. 2000;53:283–328. - PubMed
-
- Fersht A R. Structure and Mechanism in Protein Science. New York: Freeman; 1998.
-
- Jackson S E, Fersht A R. Biochemistry. 1991;30:10428–10435. - PubMed
-
- Schindler T, Herrler M, Marahiel M A, Schmid F X. Nat Struct Biol. 1995;2:663–673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

