A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis
- PMID: 11087857
- PMCID: PMC27172
- DOI: 10.1073/pnas.97.24.13027
A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis
Abstract
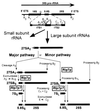
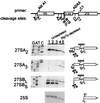
The Saccharomyces cerevisiae Rlp7 protein has extensive identity and similarity to the large ribosomal subunit L7 proteins and shares an RNA-binding domain with them. Rlp7p is not a ribosomal protein; however, it is encoded by an essential gene and therefore must perform a function essential for cell growth. In this report, we show that Rlp7p is a nucleolar protein that plays a critical role in processing of precursors to the large ribosomal subunit RNAs. Pulse-chase labeling experiments with Rlp7p-depleted cells reveal that neither 5.8S(S), 5.8S(L), nor 25S is produced, indicating that both the major and minor processing pathways are affected. Analysis of processing intermediates by primer extension indicates that Rlp7p-depleted cells accumulate the 27SA(3) precursor RNA, which is normally the major substrate (85%) used to produce the 5.8S and 25S rRNAs, and the ratio of 27SB(L) to 27SB(S) precursors changes from approximately 1:8 to 8:1 (depleted cells). Because 27SA(3) is the direct precursor to 27SB(S), we conclude that Rlp7p is specifically required for the 5' to 3' exonucleolytic trimming of the 27SA(3) into the 27SB(S) precursor. As it is essential for processing in both the major and minor pathways, we propose that Rlp7p may act as a specificity factor that binds precursor rRNAs and tethers the enzymes that carry out the early 5' to 3' exonucleolytic reactions that generate the mature rRNAs. Rlp7p may also be required for the endonucleolytic cleavage in internal transcribed spacer 2 that separates the 5.8S rRNA from the 25S rRNA.
Figures



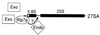
References
-
- Nissen P, Hansen J, Ban N, Moore P B, Steitz T A. Science. 2000;289:920–930. - PubMed
-
- Warner J R. Trends Biochem Sci. 1999;24:437–439. - PubMed
-
- Volarevic S, Stewart M J, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma S C, Thomas G. Science. 2000;288:2045–2047. - PubMed
-
- Venema J, Tollervey D. Annu Rev Genet. 1999;33:261–311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

