Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2
- PMID: 11087859
- PMCID: PMC27177
- DOI: 10.1073/pnas.97.24.13057
Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2
Abstract
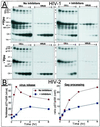
Retrovirus assembly and maturation involve folding and transport of viral proteins to the virus assembly site followed by subsequent proteolytic cleavage of the Gag polyprotein within the nascent virion. We report that inhibiting proteasomes severely decreases the budding, maturation, and infectivity of HIV. Although processing of the Env glycoproteins is not changed, proteasome inhibitors inhibit processing of Gag polyprotein by the viral protease without affecting the activity of the HIV-1 viral protease itself, as demonstrated by in vitro processing of HIV-1 Gag polyprotein Pr55. Furthermore, this effect occurs independently of the virus release function of the HIV-1 accessory protein Vpu and is not limited to HIV-1, as proteasome inhibitors also reduce virus release and Gag processing of HIV-2. Electron microscopy analysis revealed ultrastructural changes in budding virions similar to mutants in the late assembly domain of p6(gag), a C-terminal domain of Pr55 required for efficient virus maturation and release. Proteasome inhibition reduced the level of free ubiquitin in HIV-1-infected cells and prevented monoubiquitination of p6(gag). Consistent with this, viruses with mutations in PR or p6(gag) were resistant to detrimental effects mediated by proteasome inhibitors. These results indicate the requirement for an active proteasome/ubiquitin system in release and maturation of infectious HIV particles and provide a potential pharmaceutical strategy for interfering with retrovirus replication.
Figures




Comment in
-
Ubiquitin in retrovirus assembly: actor or bystander?Proc Natl Acad Sci U S A. 2000 Nov 21;97(24):12945-7. doi: 10.1073/pnas.97.24.12945. Proc Natl Acad Sci U S A. 2000. PMID: 11087848 Free PMC article. Review. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

