Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior
- PMID: 11087877
- PMCID: PMC27233
- DOI: 10.1073/pnas.97.24.13384
Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior
Abstract
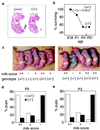
Although extracellular application of lysophosphatidic acid (LPA) has been extensively documented to produce a variety of cellular responses through a family of specific G protein-coupled receptors, the in vivo organismal role of LPA signaling remains largely unknown. The first identified LPA receptor gene, lp(A1)/vzg-1/edg-2, was previously shown to have remarkably enriched embryonic expression in the cerebral cortex and dorsal olfactory bulb and postnatal expression in myelinating glia including Schwann cells. Here, we show that targeted deletion of lp(A1) results in approximately 50% neonatal lethality, impaired suckling in neonatal pups, and loss of LPA responsivity in embryonic cerebral cortical neuroblasts with survivors showing reduced size, craniofacial dysmorphism, and increased apoptosis in sciatic nerve Schwann cells. The suckling defect was responsible for the death among lp(A1)((-/-)) neonates and the stunted growth of survivors. Impaired suckling behavior was attributable to defective olfaction, which is likely related to developmental abnormalities in olfactory bulb and/or cerebral cortex. Our results provide evidence that endogenous lysophospholipid signaling requires an lp receptor gene and indicate that LPA signaling through the LP(A1) receptor is required for normal development of an inborn, neonatal behavior.
Figures




References
-
- van Corven E J, Groenink A, Jalink K, Eichholtz T, Moolenaar W H. Cell. 1989;59:45–54. - PubMed
-
- Ridley A J, Hall A. Cell. 1992;70:389–399. - PubMed
-
- Yatomi Y, Igarashi Y, Yang L, Hisano N, Qi R, Asazuma N, Satoh K, Ozaki Y, Kume S. J Biochem (Tokyo) 1997;121:969–973. - PubMed
-
- Moolenaar W H, Kranenburg O, Postma F R, Zondag G C. Curr Opin Cell Biol. 1997;9:168–173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

