mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci
- PMID: 11090212
- PMCID: PMC150161
- DOI: 10.1105/tpc.12.11.2101
mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci
Abstract
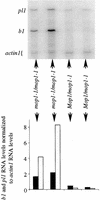
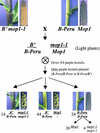
Paramutation is the directed, heritable alteration of the expression of one allele when heterozygous with another allele. Here, the isolation and characterization of a mutation affecting paramutation, mediator of paramutation1-1 (mop1-1), are described. Experiments demonstrate that the wild-type gene Mop1 is required for establishment and maintenance of the paramutant state. The mop1-1 mutation affects paramutation at the multiple loci tested but has no effect on alleles that do not participate in paramutation. The mutation does not alter the amounts of actin and ubiquitin transcripts, which suggests that the mop1 gene does not encode a global repressor. Maize plants homozygous for mop1-1 can have pleiotropic developmental defects, suggesting that mop1-1 may affect more genes than just the known paramutant ones. The mop1-1 mutation does not alter the extent of DNA methylation in rDNA and centromeric repeats. The observation that mop1 affects paramutation at multiple loci, despite major differences between these loci in their gene structure, correlations with DNA methylation, and stability of the paramutant state, suggests that a common mechanism underlies paramutation. A protein-based epigenetic model for paramutation is discussed.
Figures








References
-
- Amedeo, P., Habu, Y., Afsar, K., Mittelsten Scheid, O., and Paszkowski, J. (2000). Disruption of the plant gene MOM re-leases transcriptional silencing of methylated genes. Nature 405, 203–206. - PubMed
-
- Brink, R.A. (1958). Basis of a genetic change which invariably occurs in certain maize heterozygotes. Science 127, 1182–1183. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

