Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae
- PMID: 11094068
- PMCID: PMC102174
- DOI: 10.1128/MCB.20.24.9162-9172.2000
Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae
Abstract
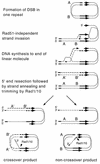
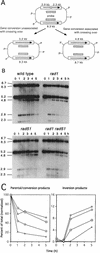
A number of studies of Saccharomyces cerevisiae have revealed RAD51-independent recombination events. These include spontaneous and double-strand break-induced recombination between repeated sequences, and capture of a chromosome arm by break-induced replication. Although recombination between inverted repeats is considered to be a conservative intramolecular event, the lack of requirement for RAD51 suggests that repair can also occur by a nonconservative mechanism. We propose a model for RAD51-independent recombination by one-ended strand invasion coupled to DNA synthesis, followed by single-strand annealing. The Rad1/Rad10 endonuclease is required to trim intermediates formed during single-strand annealing and thus was expected to be required for RAD51-independent events by this model. Double-strand break repair between plasmid-borne inverted repeats was less efficient in rad1 rad51 double mutants than in rad1 and rad51 strains. In addition, repair events were delayed and frequently associated with plasmid loss. Furthermore, the repair products recovered from the rad1 rad51 strain were primarily in the crossover configuration, inconsistent with conservative models for mitotic double-strand break repair.
Figures






Similar articles
-
Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes.Genetics. 1995 Jan;139(1):109-23. doi: 10.1093/genetics/139.1.109. Genetics. 1995. PMID: 7705617 Free PMC article.
-
RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates.Mol Cell Biol. 2000 Feb;20(4):1194-205. doi: 10.1128/MCB.20.4.1194-1205.2000. Mol Cell Biol. 2000. PMID: 10648605 Free PMC article.
-
RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae.Mol Cell Biol. 1995 Apr;15(4):2245-51. doi: 10.1128/MCB.15.4.2245. Mol Cell Biol. 1995. PMID: 7891718 Free PMC article.
-
Double-strand break repair: are Rad51/RecA--DNA joints barriers to DNA replication?Trends Genet. 2001 Jun;17(6):318-21. doi: 10.1016/s0168-9525(01)02309-5. Trends Genet. 2001. PMID: 11377793 Review.
-
DNA repair: RAD alert.Curr Biol. 1997 Aug 1;7(8):R492-5. doi: 10.1016/s0960-9822(06)00246-6. Curr Biol. 1997. PMID: 9259545 Review.
Cited by
-
BRCA2 Promotes Spontaneous Homologous Recombination In Vivo.Cancers (Basel). 2021 Jul 21;13(15):3663. doi: 10.3390/cancers13153663. Cancers (Basel). 2021. PMID: 34359565 Free PMC article.
-
Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination.EMBO J. 2001 Nov 1;20(21):6127-39. doi: 10.1093/emboj/20.21.6127. EMBO J. 2001. PMID: 11689452 Free PMC article.
-
Rad51 protein controls Rad52-mediated DNA annealing.J Biol Chem. 2008 May 23;283(21):14883-92. doi: 10.1074/jbc.M801097200. Epub 2008 Mar 12. J Biol Chem. 2008. PMID: 18337252 Free PMC article.
-
Role of Double-Strand Break End-Tethering during Gene Conversion in Saccharomyces cerevisiae.PLoS Genet. 2016 Apr 13;12(4):e1005976. doi: 10.1371/journal.pgen.1005976. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27074148 Free PMC article.
-
The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing.Genetics. 2001 Oct;159(2):515-25. doi: 10.1093/genetics/159.2.515. Genetics. 2001. PMID: 11606529 Free PMC article.
References
-
- Aguilera A. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr Genet. 1995;27:298–305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
