The MN1-TEL fusion protein, encoded by the translocation (12;22)(p13;q11) in myeloid leukemia, is a transcription factor with transforming activity
- PMID: 11094079
- PMCID: PMC102185
- DOI: 10.1128/MCB.20.24.9281-9293.2000
The MN1-TEL fusion protein, encoded by the translocation (12;22)(p13;q11) in myeloid leukemia, is a transcription factor with transforming activity
Abstract
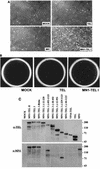
The Tel gene (or ETV6) is the target of the translocation (12;22)(p13;q11) in myeloid leukemia. TEL is a member of the ETS family of transcription factors and contains the pointed protein interaction (PNT) domain and an ETS DNA binding domain (DBD). By contrast to other chimeric proteins that contain TEL's PNT domain, such as TEL-platelet-derived growth factor beta receptor in t(5;12)(q33;p13), MN1-TEL contains the DBD of TEL. The N-terminal MN1 moiety is rich in proline residues and contains two polyglutamine stretches, suggesting that MN1-TEL may act as a deregulated transcription factor. We now show that MN1-TEL type I, unlike TEL and MN1, transforms NIH 3T3 cells. The transforming potential depends on both N-terminal MN1 sequences and a functional TEL DBD. Furthermore, we demonstrate that MN1 has transcription activity and that MN1-TEL acts as a chimeric transcription factor on the Moloney sarcoma virus long terminal repeat and a synthetic promoter containing TEL binding sites. The transactivating capacity of MN1-TEL depended on both the DBD of TEL and sequences in MN1. MN1-TEL contributes to leukemogenesis by a mechanism distinct from that of other chimeric proteins containing TEL.
Figures



Similar articles
-
MN1-TEL myeloid oncoprotein expressed in multipotent progenitors perturbs both myeloid and lymphoid growth and causes T-lymphoid tumors in mice.Blood. 2005 Dec 15;106(13):4278-86. doi: 10.1182/blood-2005-04-1674. Epub 2005 Aug 4. Blood. 2005. PMID: 16081688 Free PMC article.
-
Translocation (12;22) (p13;q11) in myeloproliferative disorders results in fusion of the ETS-like TEL gene on 12p13 to the MN1 gene on 22q11.Oncogene. 1995 Apr 20;10(8):1511-9. Oncogene. 1995. PMID: 7731705
-
MN1-TEL, the product of the t(12;22) in human myeloid leukemia, immortalizes murine myeloid cells and causes myeloid malignancy in mice.Leukemia. 2006 Sep;20(9):1582-92. doi: 10.1038/sj.leu.2404298. Epub 2006 Jun 29. Leukemia. 2006. PMID: 16810199
-
Mechanisms of transcriptional repression by the t(8;21)-, t(12;21)-, and inv(16)-encoded fusion proteins.Cancer Chemother Pharmacol. 2001 Aug;48 Suppl 1:S31-4. doi: 10.1007/s002800100302. Cancer Chemother Pharmacol. 2001. PMID: 11587363 Review.
-
Proteins of the ETS family with transcriptional repressor activity.Oncogene. 2000 Dec 18;19(55):6524-32. doi: 10.1038/sj.onc.1204045. Oncogene. 2000. PMID: 11175368 Review.
Cited by
-
Mapping of MN1 sequences necessary for myeloid transformation.PLoS One. 2013 Apr 23;8(4):e61706. doi: 10.1371/journal.pone.0061706. Print 2013. PLoS One. 2013. PMID: 23626719 Free PMC article.
-
Microdeletion del(22)(q12.2) encompassing the facial development-associated gene, MN1 (meningioma 1) in a child with Pierre-Robin sequence (including cleft palate) and neurofibromatosis 2 (NF2): a case report and review of the literature.BMC Med Genet. 2012 Mar 22;13:19. doi: 10.1186/1471-2350-13-19. BMC Med Genet. 2012. PMID: 22436304 Free PMC article. Review.
-
MN1, a novel player in human AML.Blood Cells Mol Dis. 2007 Nov-Dec;39(3):336-9. doi: 10.1016/j.bcmd.2007.06.009. Epub 2007 Aug 14. Blood Cells Mol Dis. 2007. PMID: 17698380 Free PMC article. Review.
-
Assembly of mTORC3 Involves Binding of ETV7 to Two Separate Sequences in the mTOR Kinase Domain.Int J Mol Sci. 2024 Sep 18;25(18):10042. doi: 10.3390/ijms251810042. Int J Mol Sci. 2024. PMID: 39337528 Free PMC article.
-
Evolution of the essential gene MN1 during the macroevolutionary transition toward patterning the vertebrate hindbrain.Proc Natl Acad Sci U S A. 2025 Jun 3;122(22):e2416061122. doi: 10.1073/pnas.2416061122. Epub 2025 May 27. Proc Natl Acad Sci U S A. 2025. PMID: 40424121 Free PMC article.
References
-
- Buijs A, Sherr S, van Baal S, van Bezouw S, van der Plas D, Geurts van Kessel A, Riegman P, Lekanne Deprez R, Zwarthoff E, Hagemeijer A, Grosveld G. Translocation (12;22)(p13;q11) in myeloproliferative disorders results in fusion of the ETS-like TEL gene on 12p13 to the MN1 gene on 22q11. Oncogene. 1995;10:1511–1519. - PubMed
-
- Carroll M, Tomasson M H, Barker G F, Golub T R, Gilliland D G. The TEL/platelet-derived growth factor β receptor (PDGFβR) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGFβR kinase-dependent signaling pathways. Proc Natl Acad Sci USA. 1996;93:14845–14850. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
